Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
Stockley, M.L., Ferdinand, A., Benedetti, G., Blencowe, P., Boyd, S.M., Calder, M., Charles, M.D., Edwardes, L.V., Ekwuru, T., Finch, H., Galbiati, A., Geo, L., Grande, D., Grinkevich, V., Holliday, N.D., Krajewski, W.W., MacDonald, E., Majithiya, J.B., McCarron, H., McWhirter, C.L., Patel, V., Pedder, C., Rajendra, E., Ranzani, M., Rigoreau, L.J.M., Robinson, H.M.R., Schaedler, T., Sirina, J., Smith, G.C.M., Swarbrick, M.E., Turnbull, A.P., Willis, S., Heald, R.A.(2022) J Med Chem 65: 13879-13891
- PubMed: 36200480
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01142
- Primary Citation of Related Structures:
7ZUS, 7ZX0, 7ZX1 - PubMed Abstract:
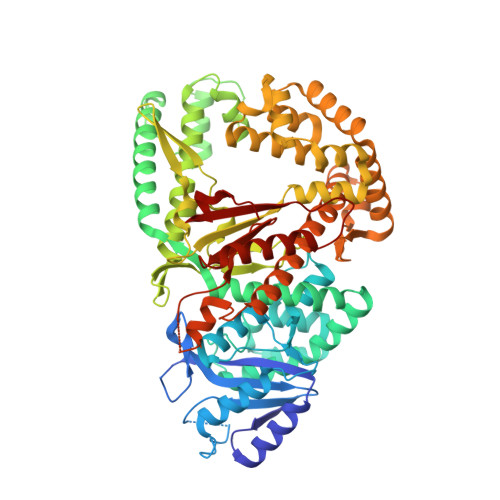
Human DNA polymerase theta (Polθ), which is essential for microhomology-mediated DNA double strand break repair, has been proposed as an attractive target for the treatment of BRCA deficient and other DNA repair pathway defective cancers. As previously reported, we recently identified the first selective small molecule Polθ in vitro probe, 22 (ART558), which recapitulates the phenotype of Polθ loss, and in vivo probe, 43 (ART812), which is efficacious in a model of PARP inhibitor resistant TNBC in vivo. Here we describe the discovery, biochemical and biophysical characterization of these probes including small molecule ligand co-crystal structures with Polθ. The crystallographic data provides a basis for understanding the unique mechanism of inhibition of these compounds which is dependent on stabilization of a "closed" enzyme conformation. Additionally, the structural biology platform provided a basis for rational optimization based primarily on reduced ligand conformational flexibility.
- Artios Pharma Ltd., B940, Babraham Research Campus, CambridgeCB22 3FH, U. K.
Organizational Affiliation: