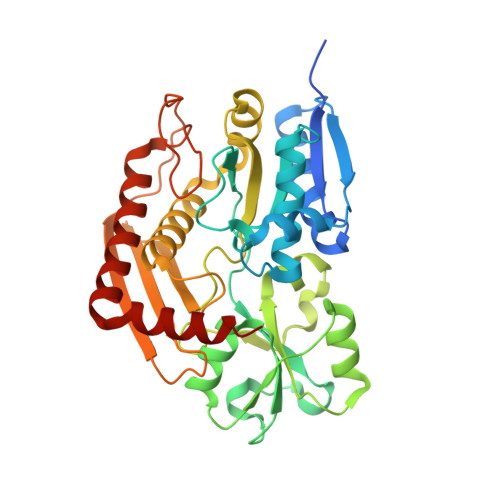
Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Bustad, H.J., Kallio, J.P., Laitaoja, M., Toska, K., Kursula, I., Martinez, A., Janis, J.(2021) iScience 24: 102152-102152
- PubMed: 33665570
- DOI: https://doi.org/10.1016/j.isci.2021.102152
- Primary Citation of Related Structures:
7AAJ, 7AAK - PubMed Abstract:
Porphobilinogen deaminase (PBGD), the third enzyme in the heme biosynthesis, catalyzes the sequential coupling of four porphobilinogen (PBG) molecules into a heme precursor. Mutations in PBGD are associated with acute intermittent porphyria (AIP), a rare metabolic disorder. We used Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) to demonstrate that wild-type PBGD and AIP-associated mutant R167W both existed as holoenzymes (E holo ) covalently attached to the dipyrromethane cofactor, and three intermediate complexes, ES, ES 2 , and ES 3 , where S represents PBG. In contrast, only ES 2 was detected in AIP-associated mutant R173W, indicating that the formation of ES 3 is inhibited. The R173W crystal structure in the ES 2 -state revealed major rearrangements of the loops around the active site, compared to wild-type PBGD in the E holo -state. These results contribute to elucidating the structural pathogenesis of two common AIP-associated mutations and reveal the important structural role of Arg173 in the polypyrrole elongation mechanism.
- Department of Biomedicine, University of Bergen, Jonas Lies vei 91, 5009 Bergen, Norway.
Organizational Affiliation: