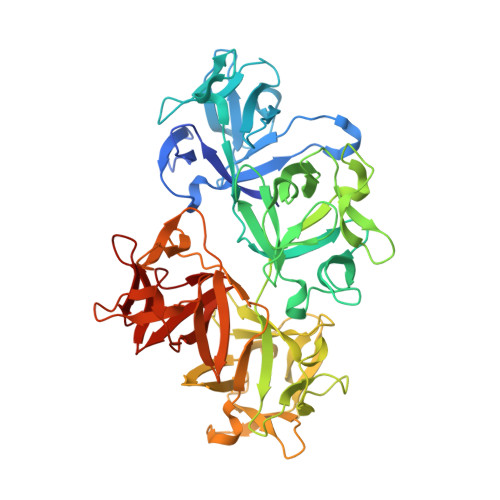
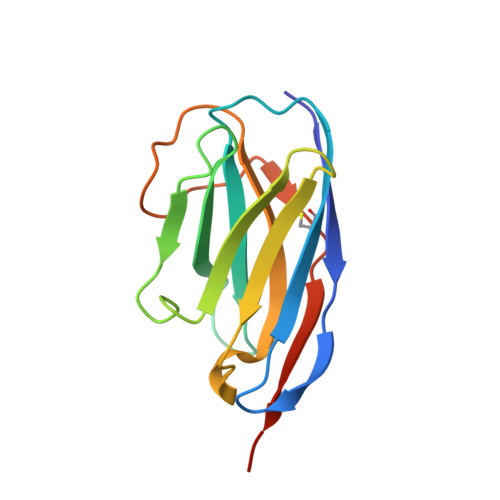
A nanobody inhibitor of Fascin-1 actin-bundling activity and filopodia formation.
Burgess, S.G., Paul, N.R., Richards, M.W., Ault, J.R., Askenatzis, L., Claydon, S.G., Corbyn, R., Machesky, L.M., Bayliss, R.(2024) Open Biol 14: 230376-230376
- PubMed: 38503329
- DOI: https://doi.org/10.1098/rsob.230376
- Primary Citation of Related Structures:
7ZAU - PubMed Abstract:
Fascin-1-mediated actin-bundling activity is central to the generation of plasma membrane protrusions required for cell migration. Dysregulated formation of cellular protrusions is observed in metastatic cancers, where they are required for increased invasiveness, and is often correlated with increased Fascin-1 abundance. Therefore, there is interest in generating therapeutic Fascin-1 inhibitors. We present the identification of Nb 3E11, a nanobody inhibitor of Fascin-1 actin-bundling activity and filopodia formation. The crystal structure of the Fascin-1/Nb 3E11 complex reveals the structural mechanism of inhibition. Nb 3E11 occludes an actin-binding site on the third β-trefoil domain of Fascin-1 that is currently not targeted by chemical inhibitors. Binding of Nb 3E11 to Fascin-1 induces a conformational change in the adjacent domains to stabilize Fascin-1 in an inhibitory state similar to that adopted in the presence of small-molecule inhibitors. Nb 3E11 could be used as a tool inhibitor molecule to aid in the development of Fascin-1 targeted therapeutics.
- Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, UK.
Organizational Affiliation: