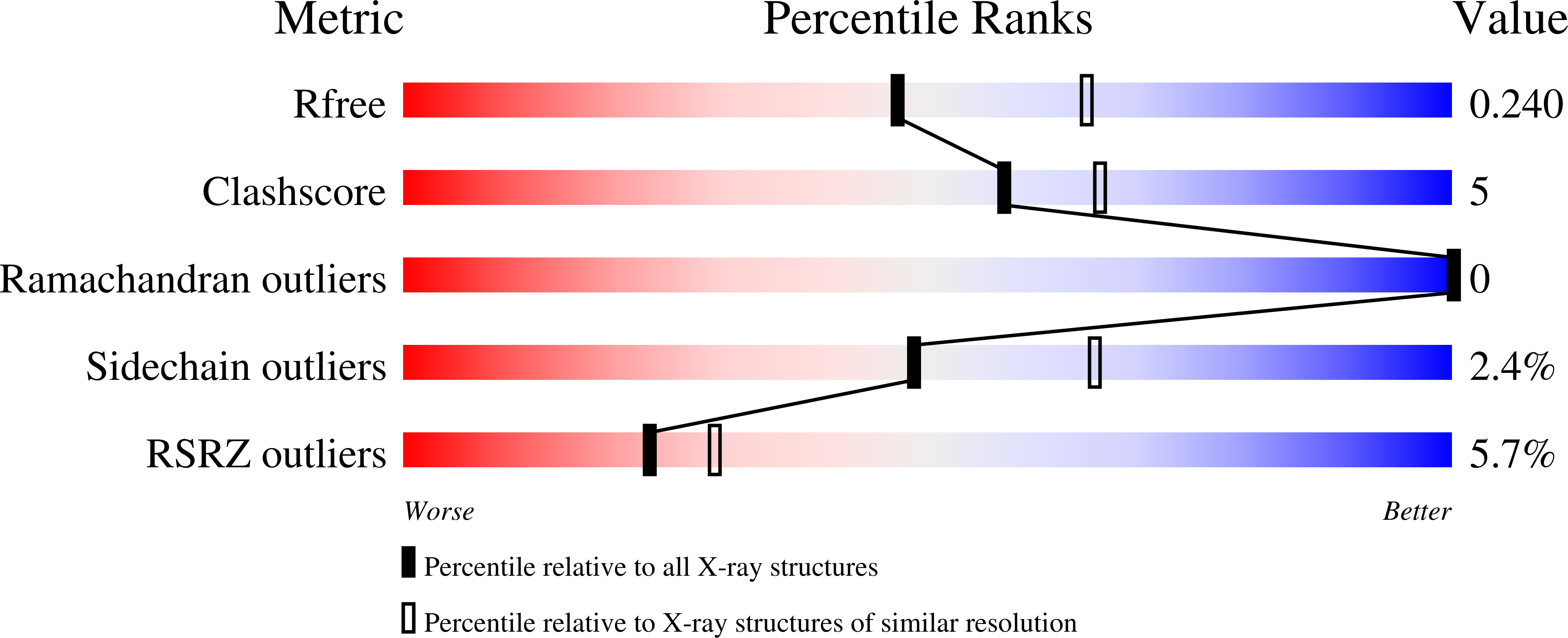
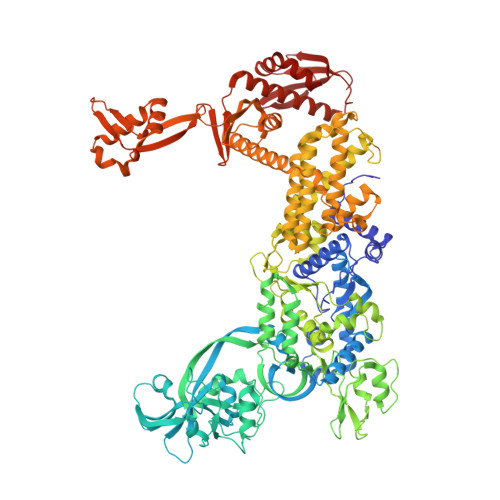
Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Brkic, A., Leibundgut, M., Jablonska, J., Zanki, V., Car, Z., Petrovic Perokovic, V., Marsavelski, A., Ban, N., Gruic-Sovulj, I.(2023) Nat Commun 14: 5498-5498
- PubMed: 37679387
- DOI: https://doi.org/10.1038/s41467-023-41244-3
- Primary Citation of Related Structures:
8C8U, 8C8V, 8C8W, 8C9D, 8C9E, 8C9F, 8C9G - PubMed Abstract:
Antibiotics target key biological processes that include protein synthesis. Bacteria respond by developing resistance, which increases rapidly due to antibiotics overuse. Mupirocin, a clinically used natural antibiotic, inhibits isoleucyl-tRNA synthetase (IleRS), an enzyme that links isoleucine to its tRNA Ile for protein synthesis. Two IleRSs, mupirocin-sensitive IleRS1 and resistant IleRS2, coexist in bacteria. The latter may also be found in resistant Staphylococcus aureus clinical isolates. Here, we describe the structural basis of mupirocin resistance and unravel a mechanism of hyper-resistance evolved by some IleRS2 proteins. We surprisingly find that an up to 10 3 -fold increase in resistance originates from alteration of the HIGH motif, a signature motif of the class I aminoacyl-tRNA synthetases to which IleRSs belong. The structural analysis demonstrates how an altered HIGH motif could be adopted in IleRS2 but not IleRS1, providing insight into an elegant mechanism for coevolution of the key catalytic motif and associated antibiotic resistance.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, University of Zagreb, Horvatovac 102a, 10000, Zagreb, Croatia.