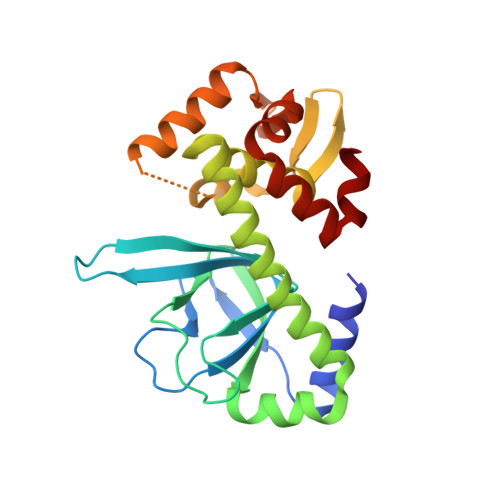
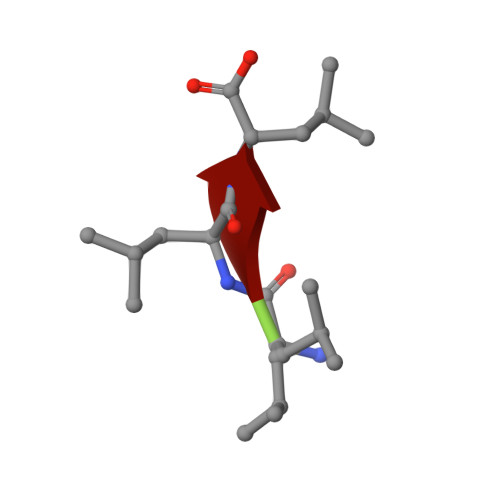
Structural basis of promiscuous inhibition of Listeria virulence activator PrfA by oligopeptides.
Hainzl, T., Scortti, M., Lindgren, C., Grundstrom, C., Krypotou, E., Vazquez-Boland, J.A., Sauer-Eriksson, A.E.(2025) Cell Rep 44: 115290-115290
- PubMed: 39970044
- DOI: https://doi.org/10.1016/j.celrep.2025.115290
- Primary Citation of Related Structures:
8CB4, 8CB5, 8CB7, 8CB8, 8CBG, 8CBI, 8CBP - PubMed Abstract:
The facultative pathogen Listeria monocytogenes uses a master regulator, PrfA, to tightly control the fitness-costly expression of its virulence factors. We found that PrfA activity is repressed via competitive occupancy of the binding site for the PrfA-activating cofactor, glutathione, by exogenous nutritional oligopeptides. The inhibitory peptides show different sequence and physicochemical properties, but how such a wide variety of oligopeptides can bind PrfA was unclear. Using crystal structure analysis of PrfA complexed with inhibitory tri- and tetrapeptides, we show here that the binding promiscuity is due to the ability of PrfA β5 in the glutathione-binding inter-domain tunnel to establish parallel or antiparallel β sheet-like interactions with the peptide backbone. Spacious tunnel pockets provide additional flexibility for unspecific peptide accommodation while providing selectivity for hydrophobic residues. Hydrophobic contributions from two adjacent peptide residues appear to be critical for efficient PrfA inhibitory binding. In contrast to glutathione, peptide binding prevents the conformational change required for the correct positioning of the DNA-binding helix-turn-helix motifs of PrfA, effectively inhibiting virulence gene expression.
Organizational Affiliation:
Department of Chemistry and Umeå Centre for Microbial Research, Umeå University, 901 87 Umeå, Sweden.