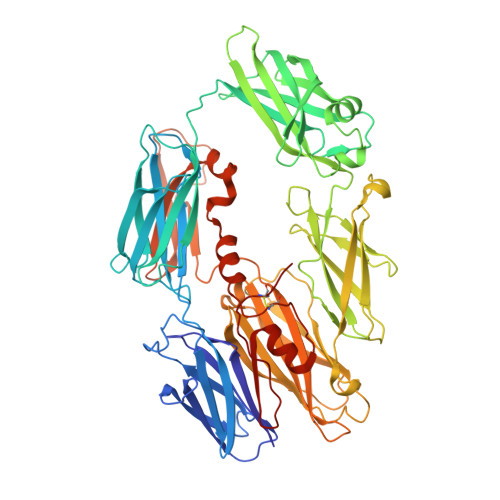
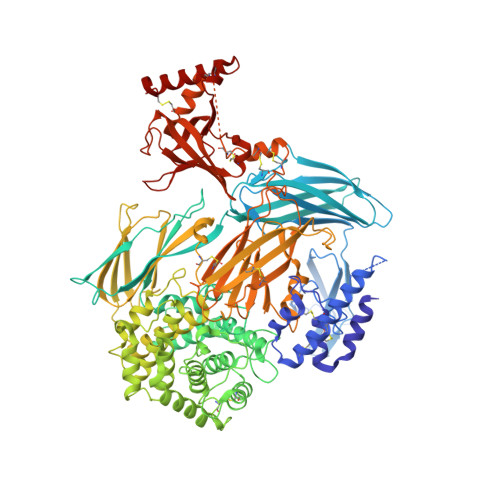
The structure of bovine complement component 3 reveals the basis for thioester function.
Fredslund, F., Jenner, L., Husted, L.B., Nyborg, J., Andersen, G.R., Sottrup-Jensen, L.(2006) J Mol Biology 361: 115-127
- PubMed: 16831446
- DOI: https://doi.org/10.1016/j.jmb.2006.06.009
- Primary Citation of Related Structures:
8CEM - PubMed Abstract:
The third component of complement (C3) is a 190 kDa glycoprotein essential for eliciting the complement response. The protein consists of two polypeptide chains (alpha and beta) held together with a single disulfide bridge. The beta-chain is composed of six MG domains, one of which is shared with the alpha-chain. The disulfide bridge connecting the chains is positioned in the shared MG domain. The alpha-chain consists of the anaphylatoxin domain, three MG domains, a CUB domain, an alpha(6)/alpha(6)-barrel domain and the C-terminal C345c domain. An internal thioester in the alpha-chain of C3 (present in C4 but not in C5) is cleaved during complement activation. This mediates covalent attachment of the activated C3b to immune complexes and invading microorganisms, thereby opsonizing the target. We present the structure of bovine C3 determined at 3 Angstroms resolution. The structure shows that the ester is buried deeply between the thioester domain and the properdin binding domain, in agreement with the human structure. This domain interface is broken upon activation, allowing nucleophile access. The structure of bovine C3 clearly demonstrates that the main chain around the thioester undergoes a helical transition upon activation. This rearrangement is proposed to be the basis for the high level of reactivity of the thioester group. A strictly conserved glutamate residue is suggested to function catalytically in thioester proteins. Structure-based design of inhibitors of C3 activation may target a conserved pocket between the alpha-chain and the beta-chain of C3, which appears essential for conformational changes in C3.
- Department of Molecular Biology, University of Aarhus, Denmark.
Organizational Affiliation: