Crystal Structure of Bifunctional protein GlmU from Klebsiella pneumoniae subsp. pneumoniae
DeBouver, N.D., Abendroth, J., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bifunctional UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase GlmU | 464 | Klebsiella pneumoniae subsp. pneumoniae | Mutation(s): 0 Gene Names: glmU, IM758_25600, KW550_14685, KW551_26820, KW555_12970, KW557_26470, KW558_26490, KW559_26805, KW560_26800, KW563_26020... | 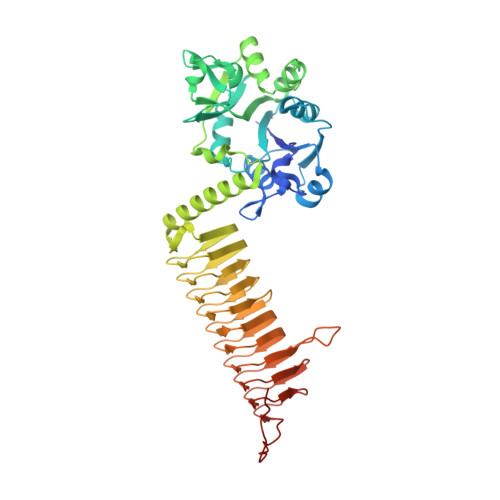 | |
UniProt | |||||
Find proteins for A6TG34 (Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)) Explore A6TG34 Go to UniProtKB: A6TG34 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A6TG34 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CIT Query on CIT | B [auth A], D [auth A] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| CL Query on CL | C [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 139.15 | α = 90 |
| b = 139.15 | β = 90 |
| c = 139.15 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| MoRDa | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |