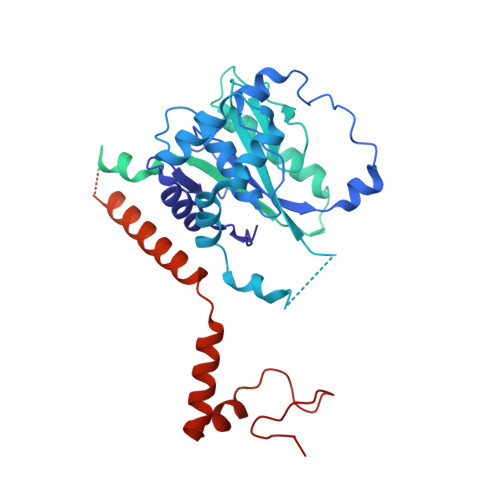
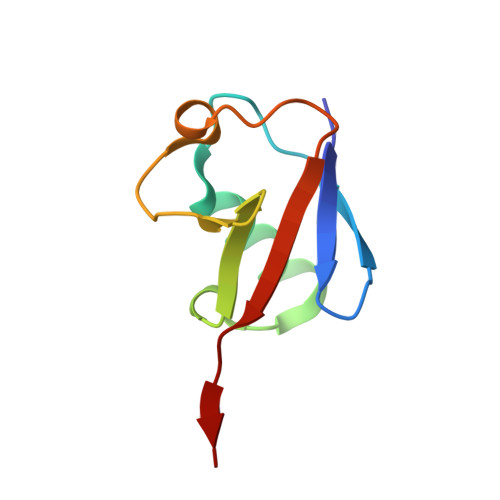
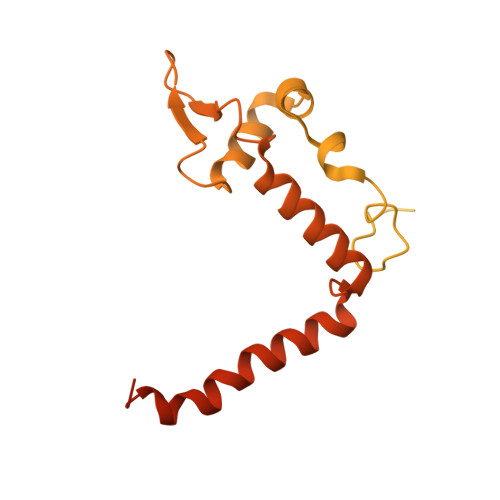
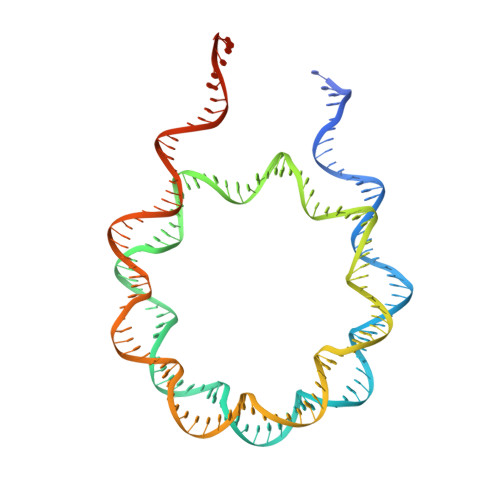
Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Ge, W., Yu, C., Li, J., Yu, Z., Li, X., Zhang, Y., Liu, C.P., Li, Y., Tian, C., Zhang, X., Li, G., Zhu, B., Xu, R.M.(2023) Nature 616: 176-182
- PubMed: 36991118
- DOI: https://doi.org/10.1038/s41586-023-05841-y
- Primary Citation of Related Structures:
8H1T - PubMed Abstract:
Repression of gene expression by protein complexes of the Polycomb group is a fundamental mechanism that governs embryonic development and cell-type specification 1-3 . The Polycomb repressive deubiquitinase (PR-DUB) complex removes the ubiquitin moiety from monoubiquitinated histone H2A K119 (H2AK119ub1) on the nucleosome 4 , counteracting the ubiquitin E3 ligase activity of Polycomb repressive complex 1 (PRC1) 5 to facilitate the correct silencing of genes by Polycomb proteins and safeguard active genes from inadvertent silencing by PRC1 (refs. 6-9 ). The intricate biological function of PR-DUB requires accurate targeting of H2AK119ub1, but PR-DUB can deubiquitinate monoubiquitinated free histones and peptide substrates indiscriminately; the basis for its exquisite nucleosome-dependent substrate specificity therefore remains unclear. Here we report the cryo-electron microscopy structure of human PR-DUB, composed of BAP1 and ASXL1, in complex with the chromatosome. We find that ASXL1 directs the binding of the positively charged C-terminal extension of BAP1 to nucleosomal DNA and histones H3-H4 near the dyad, an addition to its role in forming the ubiquitin-binding cleft. Furthermore, a conserved loop segment of the catalytic domain of BAP1 is situated near the H2A-H2B acidic patch. This distinct nucleosome-binding mode displaces the C-terminal tail of H2A from the nucleosome surface, and endows PR-DUB with the specificity for H2AK119ub1.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.