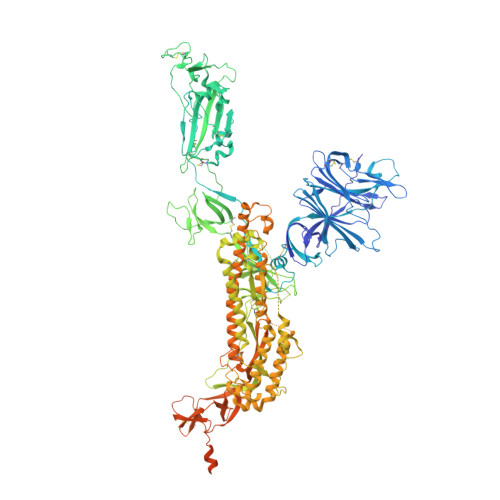
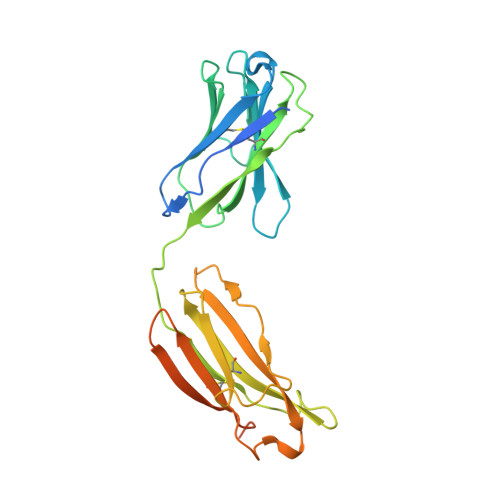
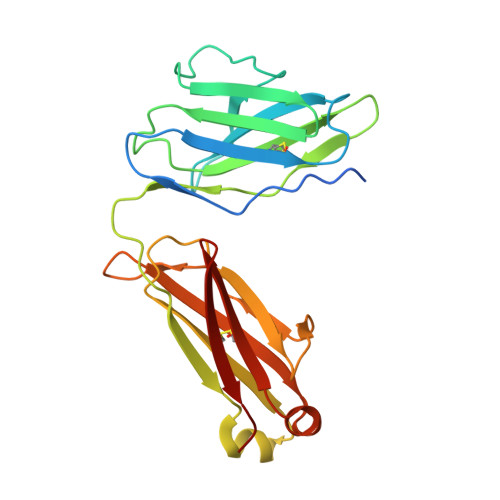
Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
Zhang, Z., Zhang, Y., Zhang, Y., Cheng, L., Zhang, L., Yan, Q., Liu, X., Chen, J., Dai, J., Guo, Y., Wei, P., Xiong, X., Xiao, J., Zhu, A., Zhuo, J., Cai, R., Zhang, J., Rao, H., Qu, B., Zhang, S., Feng, J., Cheng, J., Su, J., Chen, C., Li, S., Zhang, Y., Chen, L., Jin, Y., Xu, Y., Liu, X., Li, Y., Zhao, J., Wang, Y., Zhou, Q., Zhao, J.(2024) MedComm (2020) 5: e70008-e70008
- PubMed: 39619228
- DOI: https://doi.org/10.1002/mco2.70008
- Primary Citation of Related Structures:
8HLC, 8HLD - PubMed Abstract:
Research on virus/receptor interactions has uncovered various mechanisms of antibody-mediated neutralization against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, understanding of neutralization by antibodies targeting the silent face, which recognize epitopes on glycan shields, remains limited, and their potential protective efficacy in vivo is not well understood. This study describes a silent face neutralizing antibody, 3711, which targets a non-supersite on the N-terminal domain (NTD) of the spike protein. Cryo-EM structure determination of the 3711 Fab in the spike complex reveals a novel neutralizing epitope shielded by glycans on the spike's silent face. Antibody 3711 inhibits the interaction between the receptor-binding domain (RBD) and human angiotensin-converting enzyme 2 (hACE2) through steric hindrance and exhibits superior in vivo protective effects compared to other reported NTD-targeted monoclonal antibodies (mAbs). Competition assays and antibody repertoire analysis indicate the rarity of antibodies targeting the 3711-related epitope in SARS-CoV-2 convalescents, suggesting the infrequency of NTD silent face-targeted neutralizing antibodies during SARS-CoV-2 infection. As the first NTD silent face-targeted neutralizing antibody against SARS-CoV-2, the identification of mAb 3711, with its novel neutralizing mechanism, enhances our understanding of neutralizing epitopes on glycan shields and elucidates epitope-guided viral mutations that evade specific antibodies.
- State Key Laboratory of Respiratory Disease National Clinical Research Center for Respiratory Disease Guangzhou Institute of Respiratory Health the First Affiliated Hospital of Guangzhou Medical University Guangzhou China.
Organizational Affiliation: