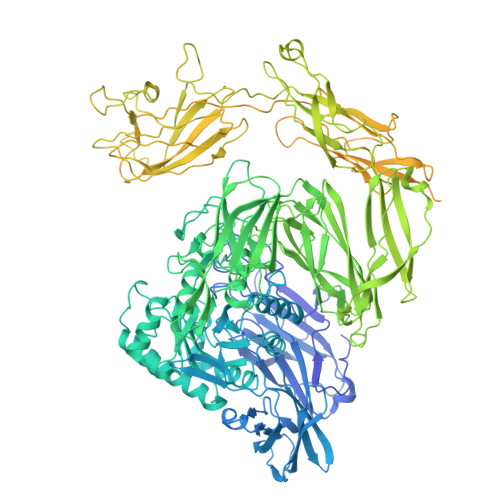
The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Hovorkova, M., Kascakova, B., Petraskova, L., Havlickova, P., Novacek, J., Pinkas, D., Gardian, Z., Kren, V., Bojarova, P., Smatanova, I.K.(2024) Structure
- PubMed: 39353423
- DOI: https://doi.org/10.1016/j.str.2024.09.005
- Primary Citation of Related Structures:
8Q2H, 8Q4Y, 8Q51 - PubMed Abstract:
β-Galactosidase from Bacillus circulans ATCC 31382 (BgaD) is a biotechnologically important enzyme for the synthesis of β-galactooligosaccharides (GOS). Among its four isoforms, isoform A (BgaD-A) has distinct synthetic properties. Here, we present cryoelectron microscopy (cryo-EM) structures of BgaD-A and compare them with the known X-ray crystal structure of isoform D (BgaD-D), revealing substantial structural divergences between the two isoforms. In contrast to BgaD-D, BgaD-A features a flexible Big-4 domain and another enigmatic domain. The newly identified flexible region in BgaD-A is termed as "barrier domain 8," and serves as a barricade, obstructing the access of longer oligosaccharide substrates into the active site of BgaD-A. The transgalactosylation reactions catalyzed by both isoforms revealed that BgaD-A has a higher selectivity than BgaD-D in the earlier stages of the reaction and is prevailingly directed to shorter galactooligosaccharides. This study improves our understanding of the structural determinants governing β-galactosidase catalysis, with implications for tailored GOS production.
Organizational Affiliation:
Laboratory of Biotransformation, Institute of Microbiology of the Czech Academy of Sciences, Vídeňská 1083, CZ-14200 Praha4, Czech Republic; Department of Genetics and Microbiology, Faculty of Science, Charles University in Prague, Viničná 5, CZ-12843 Praha2, Czech Republic.