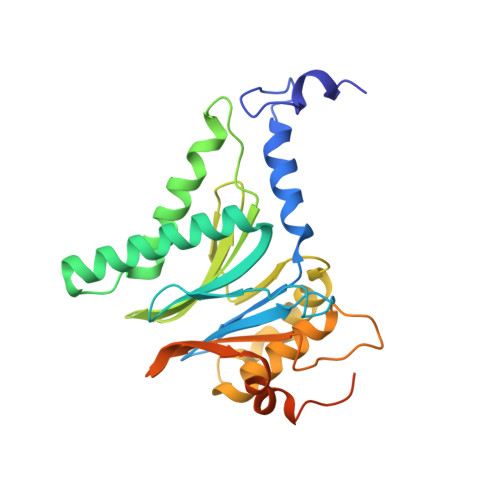
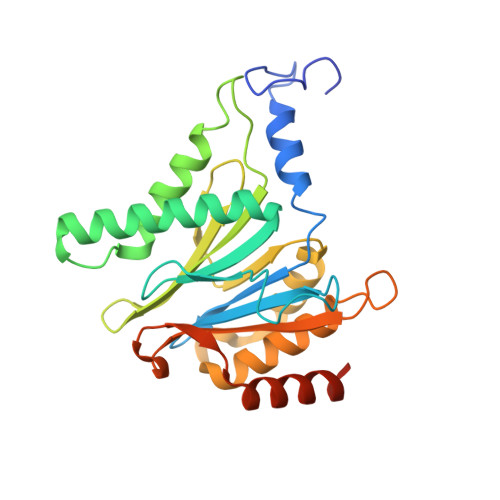
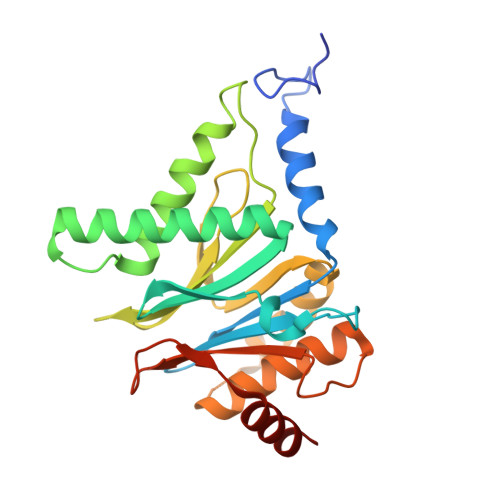
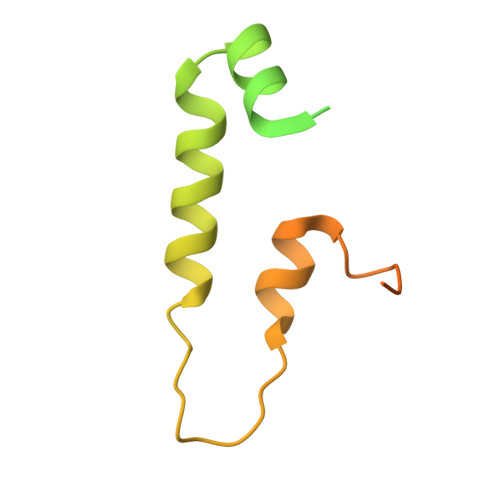
Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Adolf, F., Du, J., Goodall, E.A., Walsh Jr., R.M., Rawson, S., von Gronau, S., Harper, J.W., Hanna, J., Schulman, B.A.(2024) Nat Struct Mol Biol 31: 1176-1188
- PubMed: 38600324
- DOI: https://doi.org/10.1038/s41594-024-01268-9
- Primary Citation of Related Structures:
8QYJ, 8QYL, 8QYM, 8QYN, 8QYO, 8QYS, 8QZ9 - PubMed Abstract:
Dedicated assembly factors orchestrate the stepwise production of many molecular machines, including the 28-subunit proteasome core particle (CP) that mediates protein degradation. Here we report cryo-electron microscopy reconstructions of seven recombinant human subcomplexes that visualize all five chaperones and the three active site propeptides across a wide swath of the assembly pathway. Comparison of these chaperone-bound intermediates and a matching mature CP reveals molecular mechanisms determining the order of successive subunit additions, as well as how proteasome subcomplexes and assembly factors structurally adapt upon progressive subunit incorporation to stabilize intermediates, facilitate the formation of subsequent intermediates and ultimately rearrange to coordinate proteolytic activation with gated access to active sites. This work establishes a methodologic approach for structural analysis of multiprotein complex assembly intermediates, illuminates specific functions of assembly factors and reveals conceptual principles underlying human proteasome biogenesis, thus providing an explanation for many previous biochemical and genetic observations.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried, Germany. fadolf@biochem.mpg.de.
Organizational Affiliation: