Single particle cryo-EM co-structure of E. coli AcrB with bound BDM91531 inhibitor at 3.52 A resolution
Boernsen, C., Mueller, R.T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Multidrug efflux pump subunit AcrB | 1,049 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: acrB | 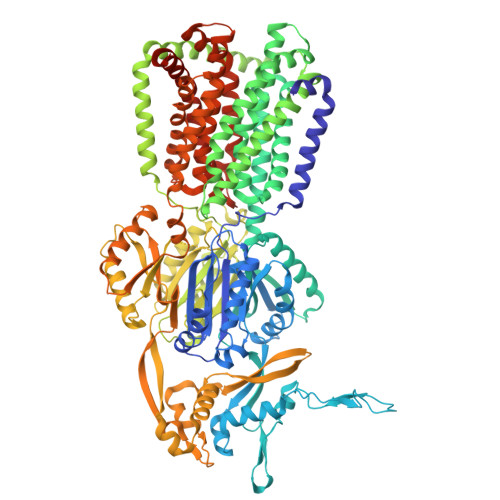 | |
UniProt | |||||
Find proteins for P31224 (Escherichia coli (strain K12)) Explore P31224 Go to UniProtKB: P31224 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31224 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XE9 (Subject of Investigation/LOI) Query on XE9 | D [auth A], E [auth C] | [3-(3-chloranyl-2-piperazin-1-yl-quinolin-6-yl)phenyl]methanamine C20 H21 Cl N4 PMBAQAZUQBDHLD-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Federal Ministry for Education and Research | Germany | BMBF- 16GW0236K |
| Agence Nationale de la Recherche (ANR) | France | ANR-19-AMRB-0007 |