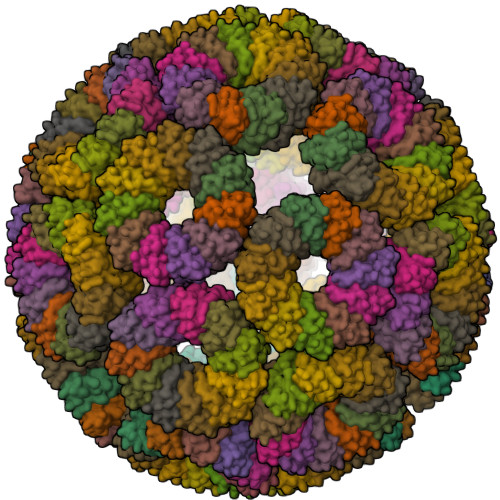
Designed, Programmable Protein Cages Utilizing Diverse Metal Coordination Geometries Show Reversible, pH-Dependent Assembly.
Osinski, N., Majsterkiewicz, K., Pakosz-Stepien, Z., Azuma, Y., Biela, A.P., Gawel, S., Heddle, J.G.(2024) Macromol Rapid Commun : e2400712-e2400712
- PubMed: 39676522
- DOI: https://doi.org/10.1002/marc.202400712
- Primary Citation of Related Structures:
8R59, 8R5A - PubMed Abstract:
The rational design and production of a novel series of engineered protein cages are presented, which have emerged as versatile and adaptable platforms with significant applications in biomedicine. These protein cages are assembled from multiple protein subunits, and precise control over their interactions is crucial for regulating assembly and disassembly, such as the on-demand release of encapsulated therapeutic agents. This approach employs a homo-undecameric, ring-shaped protein scaffold with strategically positioned metal binding sites. These engineered proteins can self-assemble into highly stable cages in the presence of cobalt or zinc ions. Furthermore, the cages can be disassembled on demand by employing external triggers such as chelating agents and changes in pH. Interestingly, for certain triggers, the disassembly process is reversible, allowing the cages to reassemble upon reversal or outcompeting of triggering conditions/agents. This work offers a promising platform for the development of advanced drug delivery systems and other biomedical applications.
Organizational Affiliation:
Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7A, Kraków, 30387, Poland.