Fragment-based development of small molecule inhibitors targeting Mycobacterium tuberculosis cholesterol metabolism.
Kavanagh, M.E., McLean, K.J., Gilbert, S.H., Amadi, C., Snee, M., Tunnicliffe, R.B., Arora, K., Boshoff, H.I., Fanourakis, A., Rebello-Lopez, M.J., Ortega-Muro, F., Levy, C.W., Munro, A.W., Leys, D., Abell, C., Coyne, A.G.(2024) bioRxiv
- PubMed: 39803573
- DOI: https://doi.org/10.1101/2024.10.28.620643
- Primary Citation of Related Structures:
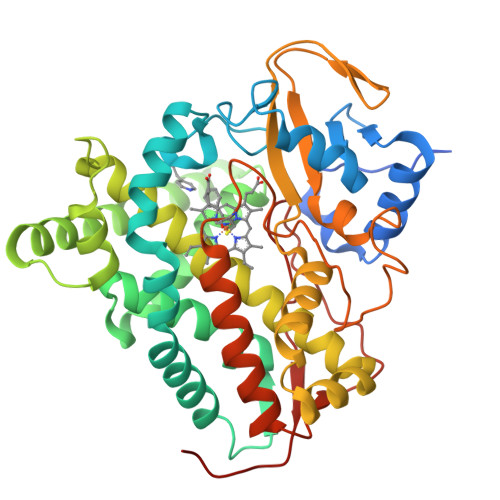
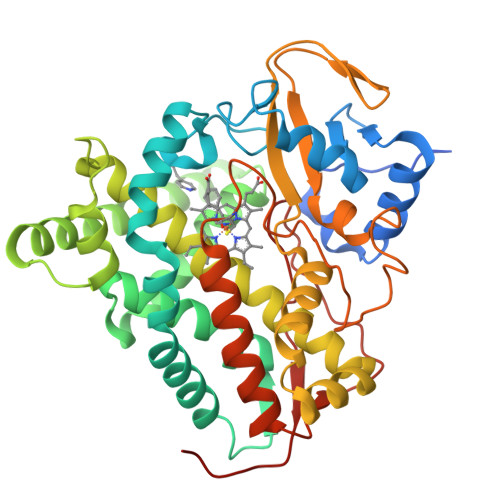
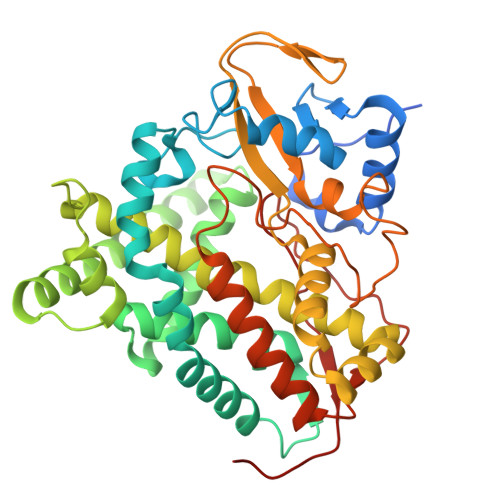
8S4M, 8S53 - PubMed Abstract:
Mycobacterium tuberculosis ( Mtb ) is the world's most deadly infectious pathogen and new drugs are urgently required to combat the emergence of multi- (MDR) and extensively- (XDR) drug resistant strains. The bacterium specifically upregulates sterol uptake pathways in infected macrophages and the metabolism of host-derived cholesterol is essential for Mtb's long-term survival in vivo. Here, we report the development of antitubercular small molecules that inhibit the Mtb cholesterol oxidases CYP125 and CYP142, which catalyze the initial step of cholesterol metabolism. An efficient biophysical fragment screen was used to characterize the structure-activity relationships of CYP125 and CYP142, and identify a non-azole small molecule 1a that can bind to the heme cofactor of both enzymes. A structure-guided fragment-linking strategy was used to optimize the binding affinity of 1a , yielding a potent dual CYP125/142 inhibitor 5m (K D CYP125/CYP142 = 0.04/0.16 μM). Compound 5m potently inhibits the catalytic activity of CYP125 and CYP142 in vitro (K I values < 0.1 μM), and rapidly depletes Mtb intracellular ATP (IC 50 = 0.15 μM). The compound has antimicrobial activity against both drug susceptible and MDR Mtb ( MIC 99 values 0.4 - 1.5 μM ) in extracellular assays, and inhibits the growth of Mtb in human macrophages (MIC = 1.7 μM) with good selectivity over mammalian cytotoxicity (LD 50 ≥ 50 μM). The combination of small molecule inhibitors and structural data reported here provide useful tools to study the role of cholesterol metabolism in Mtb and are a promising step towards novel antibiotics targeting bioenergetic pathways, which could be used to help combat MDR-TB.
Organizational Affiliation:
Yusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge, CB2 1EW, UK.