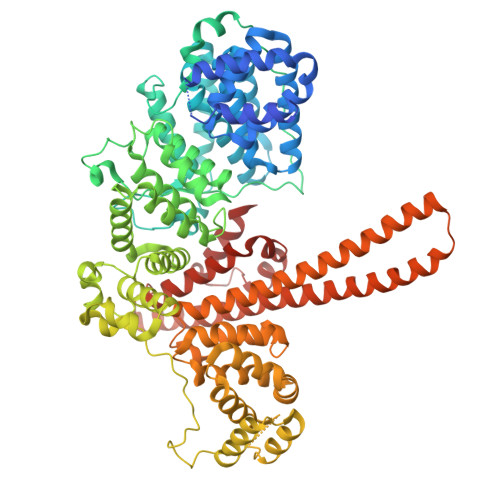
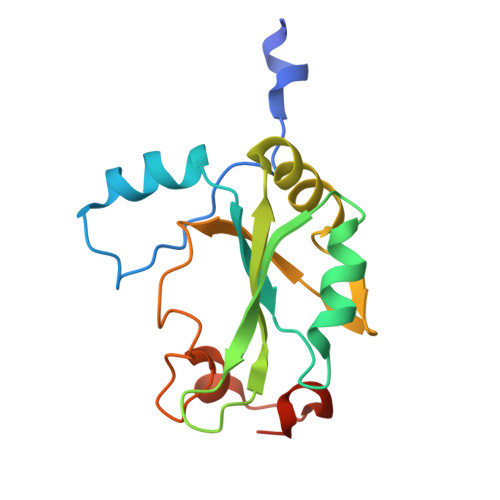
Cryo-EM structure of the CBC-ALYREF complex.
Clarke, B.P., Angelos, A.E., Mei, M., Hill, P.S., Xie, Y., Ren, Y.(2024) Elife 12
- PubMed: 39282949
- DOI: https://doi.org/10.7554/eLife.91432
- Primary Citation of Related Structures:
8SRR, 8SUY - PubMed Abstract:
In eukaryotes, RNAs transcribed by RNA Pol II are modified at the 5' end with a 7-methylguanosine (m 7 G) cap, which is recognized by the nuclear cap binding complex (CBC). The CBC plays multiple important roles in mRNA metabolism, including transcription, splicing, polyadenylation, and export. It promotes mRNA export through direct interaction with a key mRNA export factor, ALYREF, which in turn links the TRanscription and EXport (TREX) complex to the 5' end of mRNA. However, the molecular mechanism for CBC-mediated recruitment of the mRNA export machinery is not well understood. Here, we present the first structure of the CBC in complex with an mRNA export factor, ALYREF. The cryo-EM structure of CBC-ALYREF reveals that the RRM domain of ALYREF makes direct contact with both the NCBP1 and NCBP2 subunits of the CBC. Comparing CBC-ALYREF with other cellular complexes containing CBC and/or ALYREF components provides insights into the coordinated events during mRNA transcription, splicing, and export.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University School of Medicine Basic Sciences, Nashville, United States.