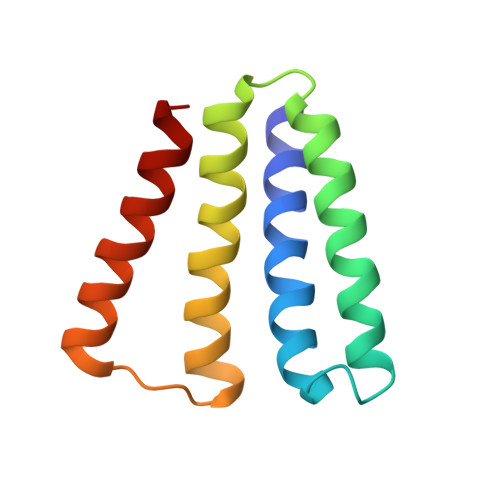
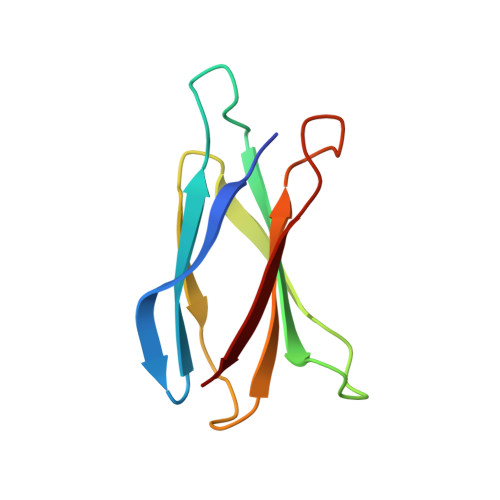
Transport of metformin metabolites by guanidinium exporters of the Small Multidrug Resistance family.
Lucero, R.M., Demirer, K., Yeh, T.J., Stockbridge, R.B.(2023) bioRxiv
- PubMed: 37645731
- DOI: https://doi.org/10.1101/2023.08.10.552832
- Primary Citation of Related Structures:
8TGY - PubMed Abstract:
Proteins from the Small Multidrug Resistance (SMR) family are frequently associated with horizontally transferred multidrug resistance gene arrays found in bacteria from wastewater and the human-adjacent biosphere. Recent studies suggest that a subset of SMR transporters might participate in metabolism of the common pharmaceutical metformin by bacterial consortia. Here, we show that both genomic and plasmid-associated transporters of the SMR Gdx functional subtype export byproducts of microbial metformin metabolism, with particularly high export efficiency for guanylurea. We use solid supported membrane electrophysiology to evaluate the transport kinetics for guanylurea and native substrate guanidinium by four representative SMR Gdx homologues. Using an internal reference to normalize independent electrophysiology experiments, we show that transport rates are comparable for genomic and plasmid-associated SMR Gdx homologues, and using a proteoliposome-based transport assay, we show that 2 proton:1 substrate transport stoichiometry is maintained. Additional characterization of guanidinium and guanylurea export properties focuses on the structurally characterized homologue, Gdx-Clo, for which we examined the pH dependence and thermodynamics of substrate binding and solved an x-ray crystal structure with guanylurea bound. Together, these experiments contribute in two main ways. By providing the first detailed kinetic examination of the structurally characterized SMR Gdx homologue Gdx-Clo, they provide a functional framework that will inform future mechanistic studies of this model transport protein. Second, this study casts light on a potential role for SMR Gdx transporters in microbial handling of metformin and its microbial metabolic byproducts, providing insight into how native transport physiologies are co-opted to contend with new selective pressures.
- Program in Chemical Biology.
Organizational Affiliation: