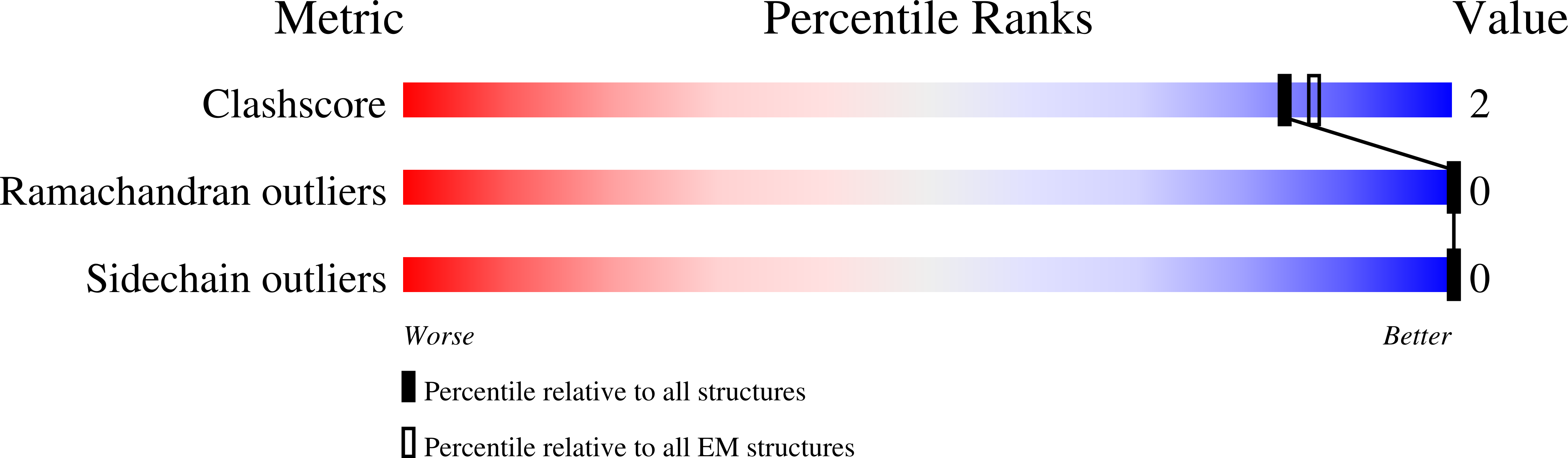Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Yang, Y.R., Han, J., Perrett, H.R., Richey, S.T., Rodriguez, A.J., Jackson, A.M., Gillespie, R.A., O'Connell, S., Raab, J.E., Cominsky, L.Y., Chopde, A., Kanekiyo, M., Houser, K.V., Chen, G.L., McDermott, A.B., Andrews, S.F., Ward, A.B.(2024) Cell Rep 43: 114171-114171
- PubMed: 38717904
- DOI: https://doi.org/10.1016/j.celrep.2024.114171
- Primary Citation of Related Structures:
8TP2, 8TP3, 8TP4, 8TP5, 8TP6, 8TP7, 8TP9, 8TPA - PubMed Abstract:
Influenza A virus subtype H2N2, which caused the 1957 influenza pandemic, remains a global threat. A recent phase 1 clinical trial investigating a ferritin nanoparticle vaccine displaying H2 hemagglutinin (HA) in H2-naive and H2-exposed adults enabled us to perform comprehensive structural and biochemical characterization of immune memory on the breadth and diversity of the polyclonal serum antibody response elicited. We temporally map the epitopes targeted by serum antibodies after vaccine prime and boost, revealing that previous H2 exposure results in higher responses to the variable HA head domain. In contrast, initial responses in H2-naive participants are dominated by antibodies targeting conserved epitopes. We use cryoelectron microscopy and monoclonal B cell isolation to describe the molecular details of cross-reactive antibodies targeting conserved epitopes on the HA head, including the receptor-binding site and a new site of vulnerability deemed the medial junction. Our findings accentuate the impact of pre-existing influenza exposure on serum antibody responses post-vaccination.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA; Chinese Academy of Sciences Key Laboratory of Nanosystem and Hierarchical Fabrication, National Center for Nanoscience and Technology, Beijing 100190, China.