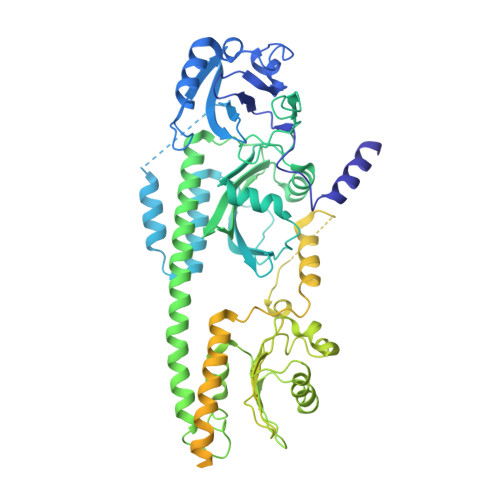
Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Burgie, E.S., Basore, K., Rau, M.J., Summers, B., Mickles, A.J., Grigura, V., Fitzpatrick, J.A.J., Vierstra, R.D.(2024) Nat Commun 15: 6853-6853
- PubMed: 39127720
- DOI: https://doi.org/10.1038/s41467-024-50412-y
- Primary Citation of Related Structures:
8U4X, 8U62, 8U63, 8U64, 8U65, 8U8Z - PubMed Abstract:
Phytochromes (Phys) are a divergent cohort of bili-proteins that detect light through reversible interconversion between dark-adapted Pr and photoactivated Pfr states. While our understandings of downstream events are emerging, it remains unclear how Phys translate light into an interpretable conformational signal. Here, we present models of both states for a dimeric Phy with histidine kinase (HK) activity from the proteobacterium Pseudomonas syringae, which were built from high-resolution cryo-EM maps (2.8-3.4-Å) of the photosensory module (PSM) and its following signaling (S) helix together with lower resolution maps for the downstream output region augmented by RoseTTAFold and AlphaFold structural predictions. The head-to-head models reveal the PSM and its photointerconversion mechanism with strong clarity, while the HK region is interpretable but relatively mobile. Pr/Pfr comparisons show that bilin phototransformation alters PSM architecture culminating in a scissoring motion of the paired S-helices linking the PSMs to the HK bidomains that ends in reorientation of the paired catalytic ATPase modules relative to the phosphoacceptor histidines. This action apparently primes autophosphorylation enroute to phosphotransfer to the cognate DNA-binding response regulator AlgB which drives quorum-sensing behavior through transient association with the photoreceptor. Collectively, these models illustrate how light absorption conformationally translates into accelerated signaling by Phy-type kinases.
Organizational Affiliation:
Department of Biology, Washington University in St. Louis, St. Louis, MO, 63130, USA.