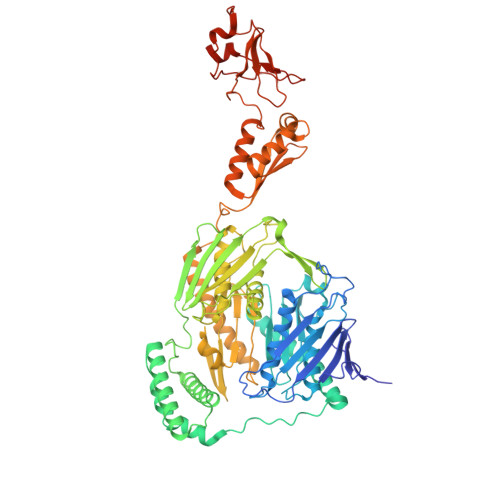
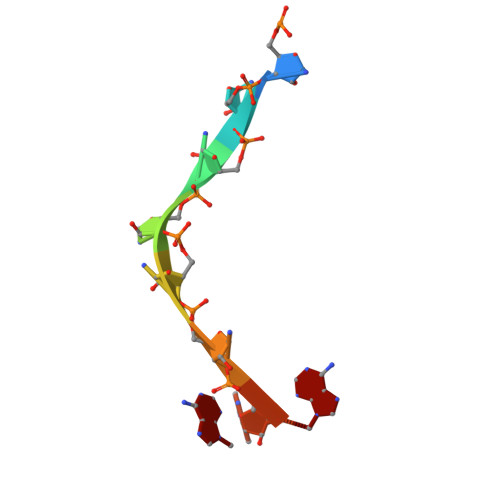
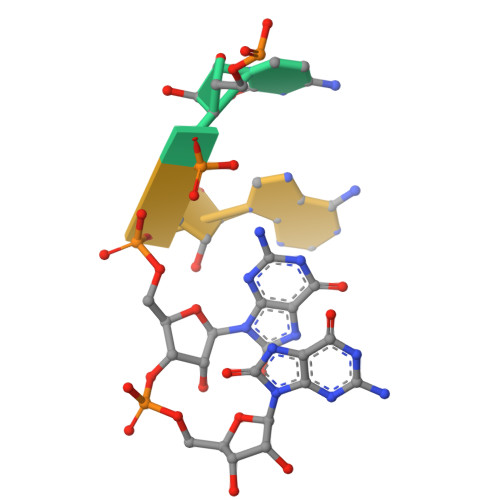
Selective 8-oxo-rG stalling occurs in the catalytic core of polynucleotide phosphorylase (PNPase) during degradation.
Miller, L.G., Kim, W., Schowe, S., Taylor, K., Han, R., Jain, V., Park, R., Sherman, M., Fang, J., Ramirez, H., Ellington, A., Tamamis, P., Resendiz, M.J.E., Zhang, Y.J., Contreras, L.(2024) Proc Natl Acad Sci U S A 121: e2317865121-e2317865121
- PubMed: 39495922
- DOI: https://doi.org/10.1073/pnas.2317865121
- Primary Citation of Related Structures:
8VAH, 8VAK - PubMed Abstract:
RNA oxidation, predominantly through the accumulation of 8-oxo-7,8-dihydroguanosine (8-oxo-rG), represents an important biomarker for cellular oxidative stress. Polynucleotide phosphorylase (PNPase) is a 3'-5' exoribonuclease that has been shown to preferentially recognize 8-oxo-rG-containing RNA and protect E scherichia coli cells from oxidative stress. However, the impact of 8-oxo-rG on PNPase-mediated RNA degradation has not been studied. Here, we show that the presence of 8-oxo-rG in RNA leads to catalytic stalling of E. coli PNPase through in vitro RNA degradation experiments and electrophoretic analysis. We also link this stalling to the active site of the enzyme through resolution of single-particle cryo-EM structures for PNPase in complex with singly or doubly oxidized RNA oligonucleotides. Following identification of Arg399 as a key residue in recognition of both single and sequential 8-oxo-rG nucleotides, we perform follow-up in vitro analysis to confirm the importance of this residue in 8-oxo-rG-specific PNPase stalling. Finally, we investigate the effects of mutations to active site residues implicated in 8-oxo-rG binding through E. coli cell growth experiments under H 2 O 2 -induced oxidative stress. Specifically, Arg399 mutations show significant effects on cell growth under oxidative stress. Overall, we demonstrate that 8-oxo-rG-specific stalling of PNPase is relevant to bacterial survival under oxidative stress and speculate that this enzyme might associate with other cellular factors to mediate this stress.
- McKetta Department of Chemical Engineering, The University of Texas at Austin, Austin, TX 78712.
Organizational Affiliation: