A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
An, T., Tan, Q., Jiang, L., Liu, L., Jiang, X., Liu, L., Chang, X., Tian, X., Deng, Z., Gao, S., Wang, L., Chen, S.(2025) Nat Chem Biol
- PubMed: 39820821
- DOI: https://doi.org/10.1038/s41589-024-01832-w
- Primary Citation of Related Structures:
8WET, 8WFD, 8Y1K - PubMed Abstract:
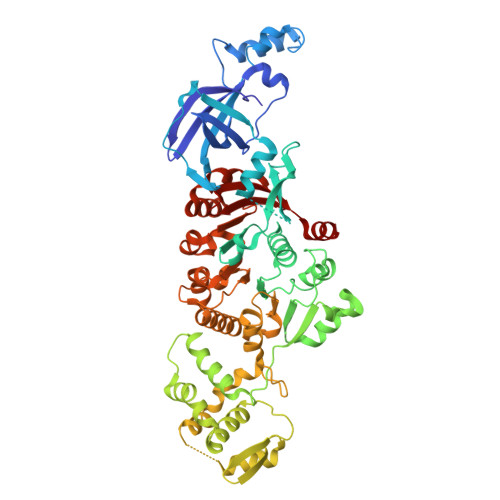
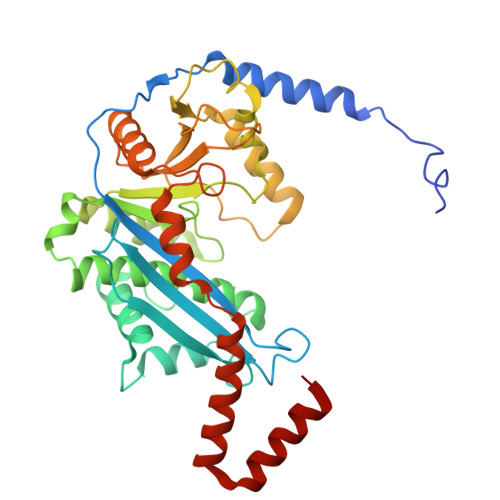
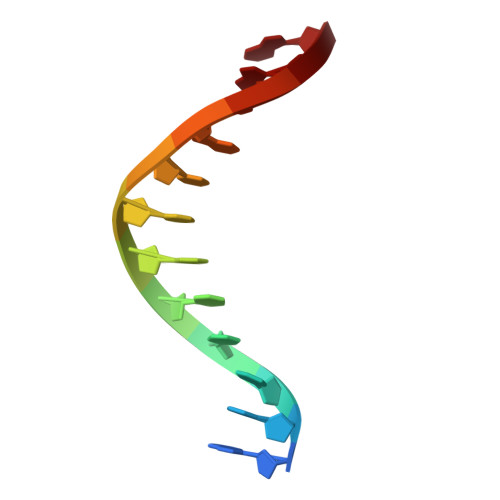
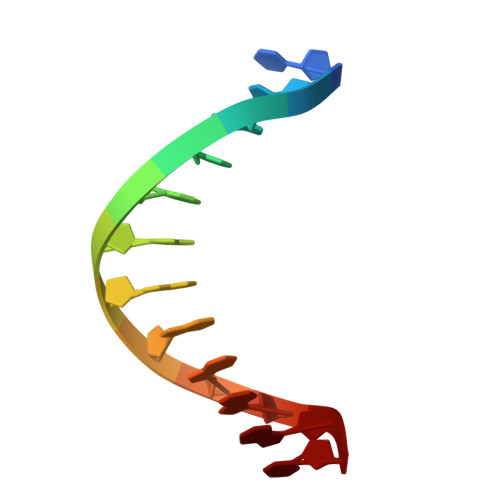
In prokaryotes, the non-bridging oxygen in the DNA sugar-phosphate backbone can be enzymatically replaced by a sulfur atom, resulting in phosphorothioate (PT) modification. However, the mechanism underlying the oxygen-to-sulfur substitution remains enigmatic. In this study, we discovered a hypercompact DNA phosphorothioation system, TdpABC, in extreme thermophiles. This DNA sulfuration process occurs through two sequential steps: an initial activation step by ATP to form an adenylated intermediate, followed by a substitution step where the adenyl group is replaced with a sulfur atom. Together with the TdpA-TdpB, the TdpABC system provides anti-phage defense by degrading PT-free phage DNA. Cryogenic electron microscopy structural analysis revealed that the TdpA hexamer binds one strand of encircled duplex DNA via hydrogen bonds arranged in a spiral staircase conformation. Nevertheless, the TdpAB-DNA interaction was sensitive to the hydrophobicity of the PT sulfur. PTs inhibit ATP-driven translocation and nuclease activity of TdpAB on self-DNA, thereby preventing autoimmunity.
Organizational Affiliation:
Department of Gastroenterology, Hubei Clinical Center and Key Laboratory of Intestinal and Colorectal Disease, TaiKang Center for Life and Medical Sciences, Zhongnan Hospital of Wuhan University, School of Pharmaceutical Sciences, Wuhan University, Wuhan, China.