structure of phage T6 topoisomerase II central domain bound with DNA
Chen, Y.T., Xin, Y.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA topoisomerase medium subunit | 452 | Escherichia phage T4 | Mutation(s): 0 Gene Names: 52 EC: 5.6.2.2 | 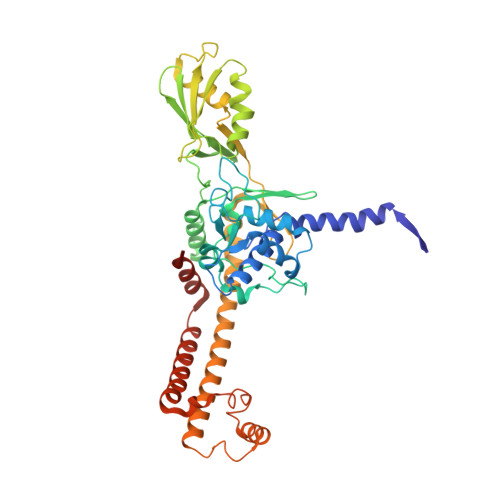 | |
UniProt | |||||
Find proteins for P07065 (Enterobacteria phage T4) Explore P07065 Go to UniProtKB: P07065 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07065 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA topoisomerase (ATP-hydrolyzing) | 611 | Salmonella phage Chi | Mutation(s): 0 Gene Names: EcT6_00003 EC: 5.6.2.2 | 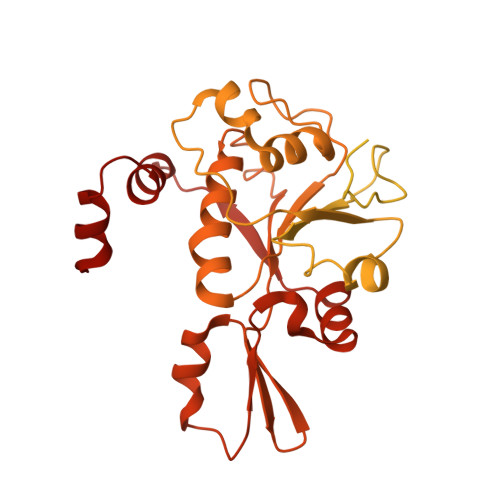 | |
UniProt | |||||
Find proteins for A0A346FJ89 (Enterobacteria phage T6) Explore A0A346FJ89 Go to UniProtKB: A0A346FJ89 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A346FJ89 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA(5'-D(P*TP*GP*TP*GP*TP*GP*TP*AP*TP*AP*TP*AP*TP*AP*CP*AP*CP*AP*TP*AP*TP*AP)) | 22 | DNA molecule | 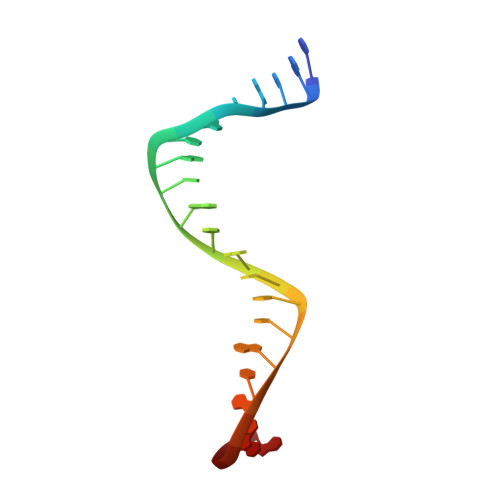 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*TP*AP*TP*AP*TP*GP*TP*GP*TP*AP*TP*AP*TP*AP*TP*AP*CP*AP*CP*AP*CP*A)-3') | 22 | DNA molecule |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG (Subject of Investigation/LOI) Query on MG | G [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | -- |