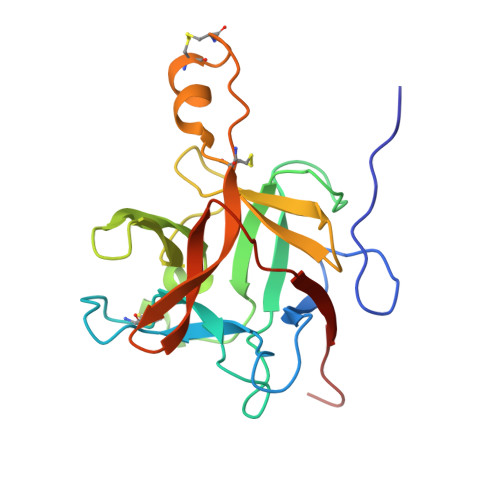
Structural determination of a new non-canonical inhibition complex between porcine trypsin and M271 a potato Kunitz-STI inhibitor.
Campuzano-Gonzalez, A., Gil-Rodriguez, P., Quintana-Armas, A.X., Guerra, Y., Perez, Y., Rudino-Pinera, E.(2025) Biochem Biophys Res Commun 768: 151818-151818
- PubMed: 40345005
- DOI: https://doi.org/10.1016/j.bbrc.2025.151818
- Primary Citation of Related Structures:
9CSZ, 9CT1 - PubMed Abstract:
The Kunitz-Soybean Trypsin Inhibitor (Kunitz-STI) is a protein family found in different plants. Several of its members have been experimentally described as inhibitors of various classes of proteases from several organisms. Even though most of the previous crystallographic studies describing the protease inhibition interactions present by different Kunitz-STI family members have been focused on serine proteases, the protein family has been proposed as a promising scaffold for multifunctional protease inhibitor design with a wide range of applications. In this work, a crystallographic complex formed between M271, a Kunitz-STI inhibitor obtained from Solanum tuberosum, and porcine trypsin shows a new inhibitory ensemble in which inhibitor's loops β1-β2 and β3-β4 play a central role. Additionally, the structural analysis demonstrates that the inhibition described here follows a non-canonical mechanism in which the inhibitor loops block the trypsin active site. Our findings expand the repertoire of protease-binding loops in which a Kunitz-STI inhibitor can inhibit trypsin, emphasizing the role of the dynamic characteristics of this protein family member on the protease inhibition process. This work also gives a new example of the remarkable plasticity of the Kunitz-STI fold, in which an arrangement with eleven highly sequence-variable loops projected to the solvent demonstrates a simple answer for a multifunctional evolutionary tool to inhibit proteases.
- Laboratorio de Bioquímica Estructural, Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México (UNAM), Morelos, Mexico. Electronic address: alexis.campuzano@ibt.unam.mx.
Organizational Affiliation: