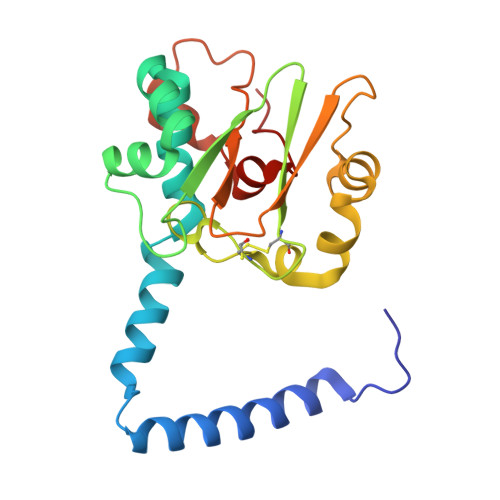
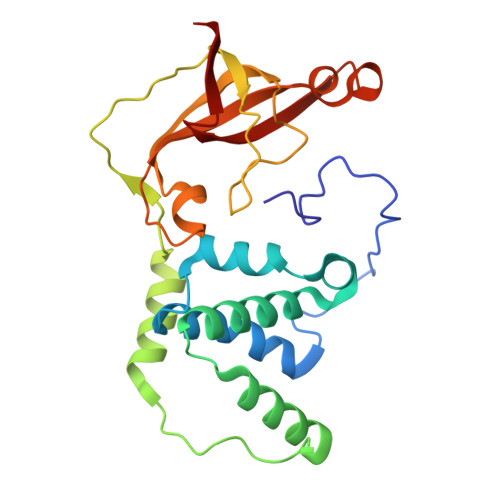
Catalytic and post-translational maturation roles of a conserved active site serine residue in nitrile hydratases.
Miller, C., Knutson, K., Liu, D., Bennett, B., Holz, R.C.(2024) J Inorg Biochem 262: 112763-112763
- PubMed: 39447484
- DOI: https://doi.org/10.1016/j.jinorgbio.2024.112763
- Primary Citation of Related Structures:
9D5U, 9D5V, 9D5Y - PubMed Abstract:
A highly conserved second-sphere active site αSer residue in nitrile hydratase (NHase), that forms a hydrogen bond with the axial metal-bound water molecule, was mutated to Ala, Asp, and Thr, in the Co-type NHase from Pseudonocardia thermophila JCM 3095 (PtNHase) and to Ala and Thr in the Fe-type NHase from Rhodococcus equi TG328-2 (ReNHase). All five mutants were successfully purified; metal analysis via ICP-AES indicated that all three Co-type PtNHase mutants were in their apo-form while the Fe-type αSer117Ala and αSer117Thr mutants contained 85 and 50 % of their active site Fe(III) ions, respectively. The k cat values obtained for the PtNHase mutant enzymes were between 0.03 ± 0.01 and 0.2 ± 0.02 s -1 amounting to <0.8 % of the k cat value observed for WT PtNHase. The Fe-type ReNHase mutants retained some detectable activity with k cat values of 93 ± 3 and 40 ± 2 s -1 for the αSer117Ala and αSer117Thr mutants, respectively, which is ∼5 % of WT ReNHase activity towards acrylonitrile. UV-Vis spectra coupled with EPR data obtained on the ReNHase mutant enzymes showed subtle changes in the electronic environment around the active site Fe(III) ions, consistent with altering the hydrogen bonding interaction with the axial water ligand. X-ray crystal structures of the three PtNHase mutant enzymes confirmed the mutation and the lack of active site metal, while also providing insight into the active site hydrogen bonding network. Taken together, these data confirm that the conserved active site αSer residue plays an important catalytic role but is not essential for catalysis. They also confirm the necessity of the conserved second-sphere αSer residue for the metalation process and subsequent post-translational modification of the α-subunit in Co-type NHases but not Fe-type NHases, suggesting different mechanisms for the two types of NHases. SYNOPSIS: A strictly conserved active site αSer residue in both Co- and Fe-type nitrile hydratases was mutated. This αSer residue was found to play an important catalytic function, but is not essential. In Co-type NHases, it appears to be essential for active site maturation, but not in Fe-type NHases.
- Department of Chemistry, Colorado School of Mines, Golden, CO 80401, USA.
Organizational Affiliation: