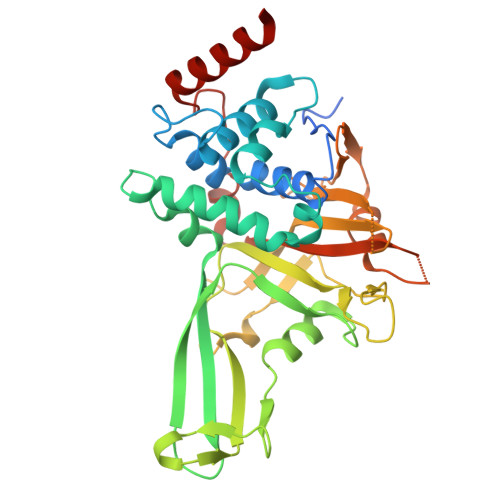
Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Miranda, R., Anson, F., Smith, S.T., Ultsch, M., Tenorio, C.A., Rouge, L., Farrell, B., Adaligil, E., Holden, J.K., Harris, S.F., Dueber, E.C.(2025) Structure 33: 705-717.e4
- PubMed: 39983720
- DOI: https://doi.org/10.1016/j.str.2025.01.021
- Primary Citation of Related Structures:
9DEK, 9DEL, 9DEM, 9DEN, 9DEO, 9DEP - PubMed Abstract:
The ubiquitin-specific protease (USP) family of deubiquitinases (DUBs) are regulators of Ub signaling that share a common catalytic-domain fold. The dynamic nature of this domain is important for controlling the function of USPs, with inter- and intramolecular interactions often influencing the structure and enzymatic activity of these DUBs. This conformational flexibility, in combination with the high sequence conservation of the USP active site, has made it challenging to readily identify potent and selective inhibitors for individual USPs. Here, we demonstrate how a naive, macrocycle-mRNA display selection rapidly yielded high-affinity binders to USP7 that specifically inhibit the DUB with nanomolar half-maximal inhibitory concentration (IC 50 ) values. Structural analysis of the macrocycles bound to USP7 revealed a variety of binding modes and identified inhibition hotspots on the enzyme that mirror those used by small-molecule inhibitors. Together, these data suggest that initial macrocyclic hits could serve as pivotal tools in developing USP-specific inhibitors and probing USP biology.
- Department of Biological Chemistry, Genentech, 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: