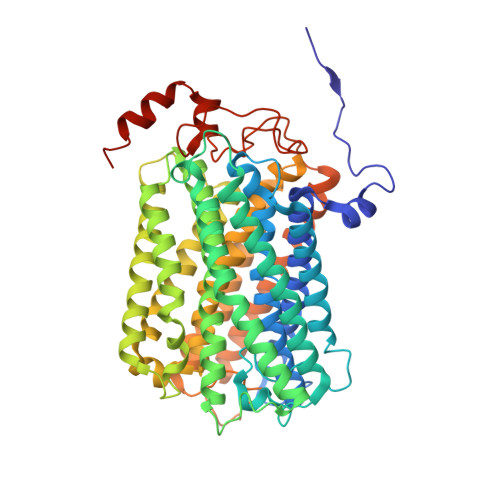
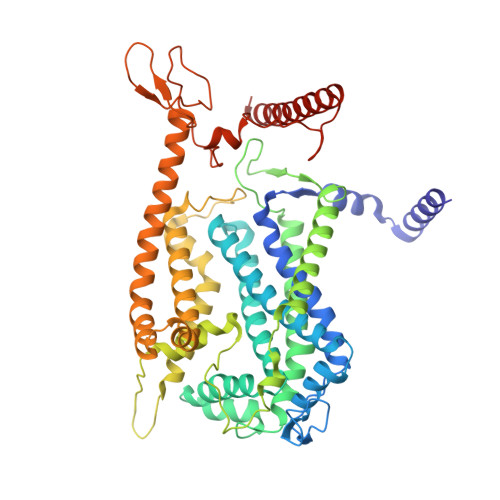
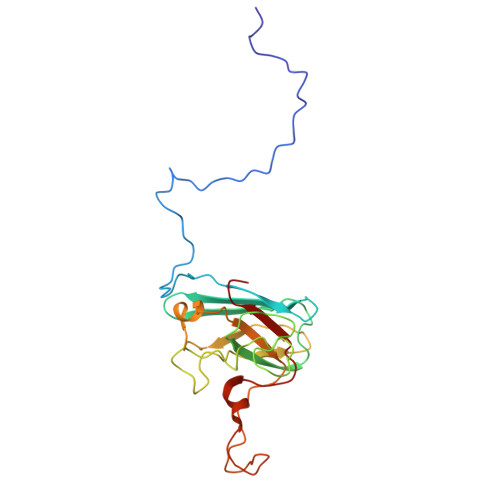
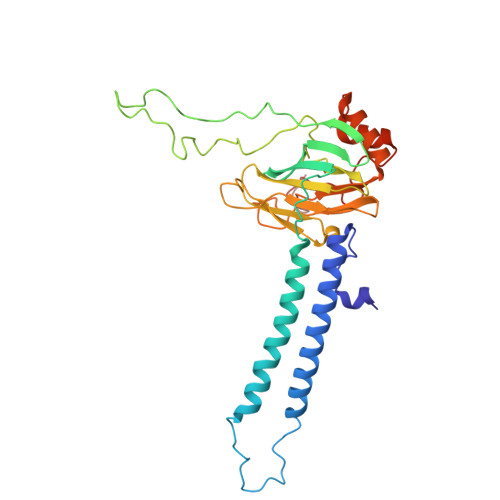
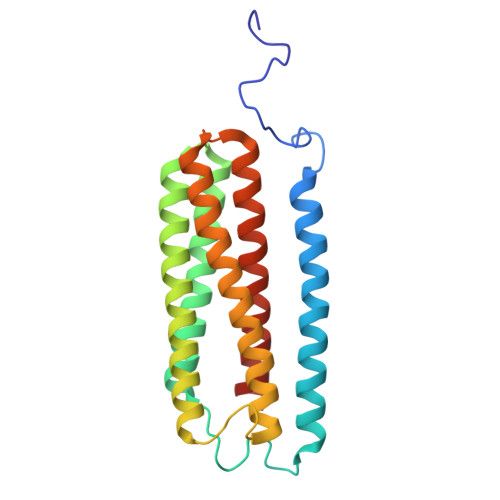
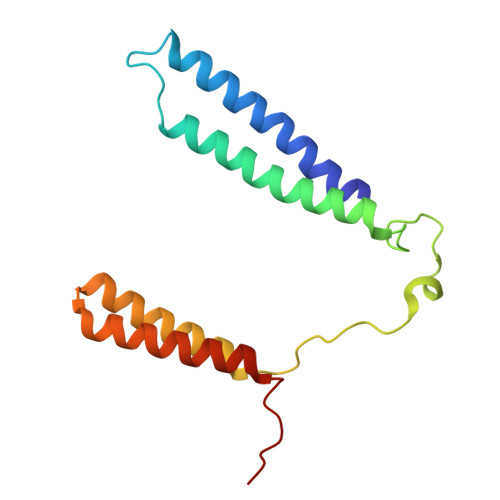
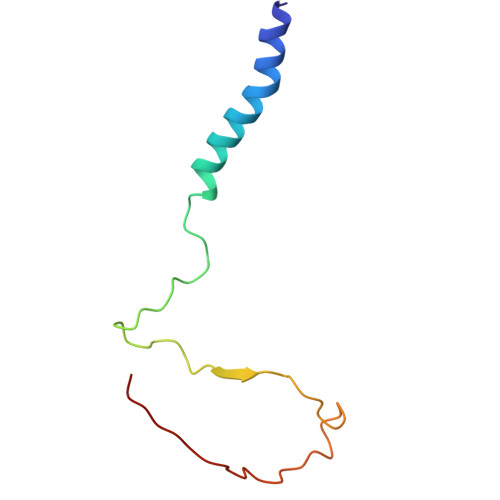
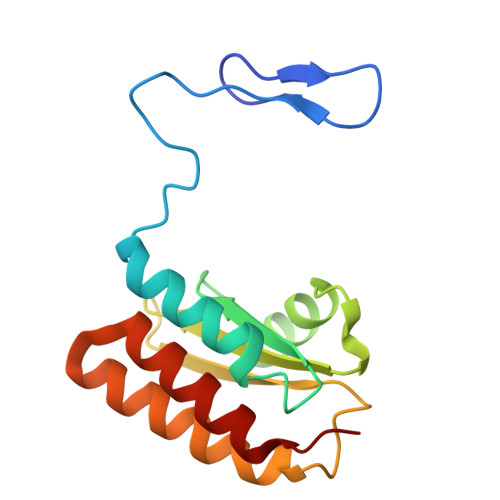
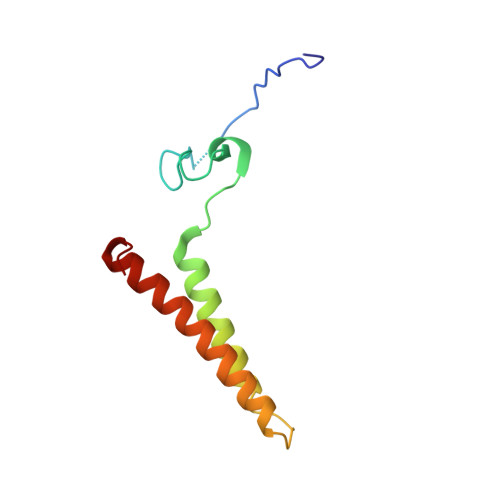
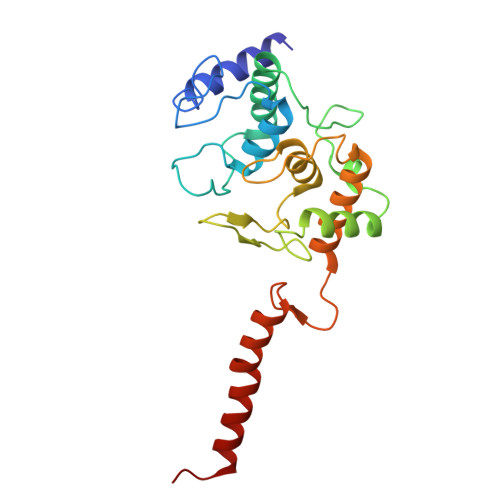
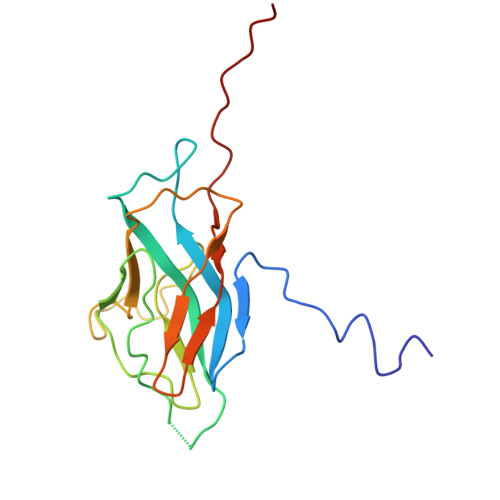
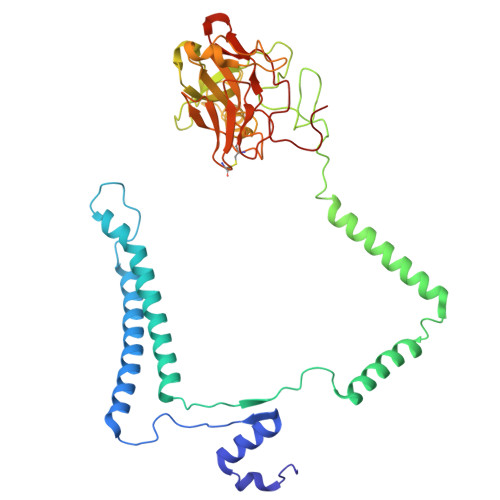
Cryo-EM of native membranes reveals an intimate connection between the Krebs cycle and aerobic respiration in mycobacteria.
Di Trani, J.M., Yu, J., Courbon, G.M., Lobez Rodriguez, A.P., Cheung, C.Y., Liang, Y., Coupland, C.E., Bueler, S.A., Cook, G.M., Brzezinski, P., Rubinstein, J.L.(2025) Proc Natl Acad Sci U S A 122: e2423761122-e2423761122
- PubMed: 39969994
- DOI: https://doi.org/10.1073/pnas.2423761122
- Primary Citation of Related Structures:
9DM1 - PubMed Abstract:
To investigate the structure of the mycobacterial oxidative phosphorylation machinery, we prepared inverted membrane vesicles from Mycobacterium smegmatis , enriched for vesicles containing complexes of interest, and imaged the vesicles with electron cryomicroscopy. We show that this analysis allows determination of the structure of both mycobacterial ATP synthase and the supercomplex of respiratory complexes III and IV in their native membrane. The latter structure reveals that the enzyme malate:quinone oxidoreductase (Mqo) physically associates with the respiratory supercomplex, an interaction that is lost on extraction of the proteins from the lipid bilayer. Mqo catalyzes an essential reaction in the Krebs cycle, and in vivo survival of mycobacterial pathogens is compromised when its activity is absent. We show with high-speed spectroscopy that the Mqo:supercomplex interaction enables rapid electron transfer from malate to the supercomplex. Further, the respiratory supercomplex is necessary for malate-driven, but not NADH-driven, electron transport chain activity and oxygen consumption. Together, these findings indicate a connection between the Krebs cycle and aerobic respiration that directs electrons along a single branch of the mycobacterial electron transport chain.
- Molecular Medicine Program, The Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.
Organizational Affiliation: