4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Peluso-Iltis, C., Pierrat, N., Rovito, D., Osz, J., Sawada, D., Kittaka, A., Laverny, G., Rochel, N.(2024) Biomolecules 14
- PubMed: 38785958
- DOI: https://doi.org/10.3390/biom14050551
- Primary Citation of Related Structures:
9EZ1, 9EZ2 - PubMed Abstract:
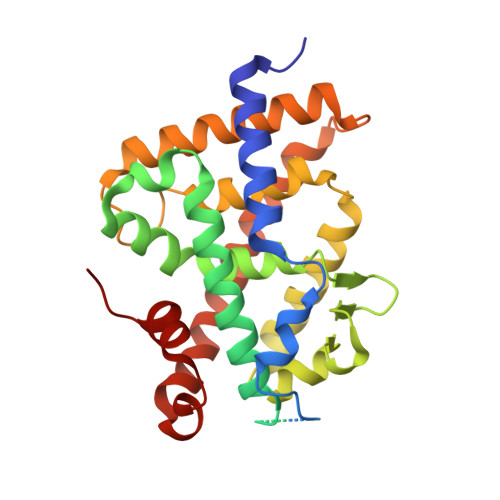
The active vitamin D metabolites, 25-hydroxyvitamin D 3 (25D 3 ) and 1,25-dihydroxyvitamin D 3 (1,25D 3 ), are produced by successive hydroxylation steps and play key roles in several cellular processes. However, alternative metabolic pathways exist, and among them, the 4-hydroxylation of 25D 3 is a major one. This study aims to investigate the structure-activity relationships of 4-hydroxy derivatives of 1,25D 3 . Structural analysis indicates that 1,4α,25(OH) 3 D 3 and 1,4β,25(OH) 3 D 3 maintain the anchoring hydrogen bonds of 1,25D 3 and form additional interactions, stabilizing the active conformation of VDR. In addition, 1,4α,25D 3 and 1,4β,25D 3 are as potent as 1,25D 3 in regulating the expression of VDR target genes in rat intestinal epithelial cells and in the mouse kidney. Moreover, these two 4-hydroxy derivatives promote hypercalcemia in mice at a dose similar to that of the parent compound.
Organizational Affiliation:
Institute of Genetics and Molecular and Cellular Biology (IGBMC), 67400 Illkirch, France.