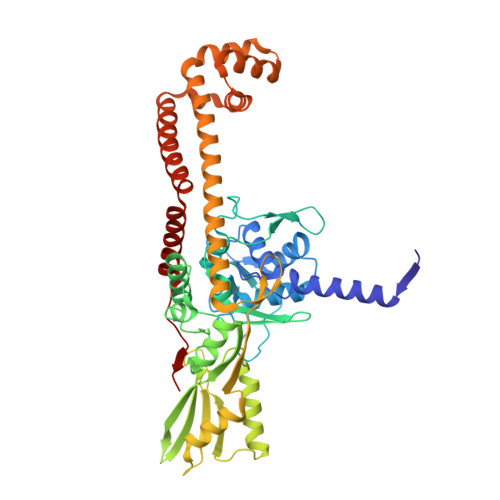
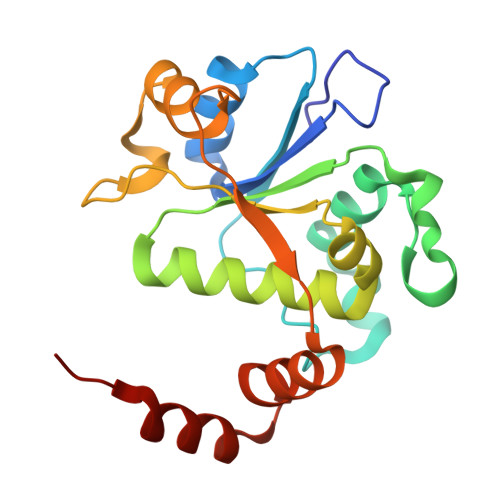
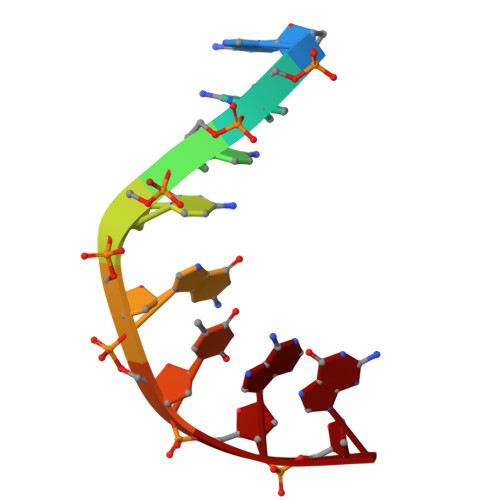
How Do Gepotidacin and Zoliflodacin Stabilize DNA Cleavage Complexes with Bacterial Type IIA Topoisomerases? 1. Experimental Definition of Metal Binding Sites.
Morgan, H., Nicholls, R.A., Warren, A.J., Ward, S.E., Evans, G., Long, F., Murshudov, G.N., Duman, R., Bax, B.D.(2024) Int J Mol Sci 25
- PubMed: 39519241
- DOI: https://doi.org/10.3390/ijms252111688
- Primary Citation of Related Structures:
9FZ6 - PubMed Abstract:
One of the challenges for experimental structural biology in the 21st century is to see chemical reactions happen. Staphylococcus aureus ( S. aureus ) DNA gyrase is a type IIA topoisomerase that can create temporary double-stranded DNA breaks to regulate DNA topology. Drugs, such as gepotidacin, zoliflodacin and the quinolone moxifloxacin, can stabilize these normally transient DNA strand breaks and kill bacteria. Crystal structures of uncleaved DNA with a gepotidacin precursor (2.1 Å GSK2999423) or with doubly cleaved DNA and zoliflodacin (or with its progenitor QPT-1) have been solved in the same P6 1 space-group (a = b ≈ 93 Å, c ≈ 412 Å). This suggests that it may be possible to observe the two DNA cleavage steps (and two DNA-religation steps) in this P6 1 space-group. Here, a 2.58 Å anomalous manganese dataset in this crystal form is solved, and four previous crystal structures (1.98 Å, 2.1 Å, 2.5 Å and 2.65 Å) in this crystal form are re-refined to clarify crystal contacts. The structures clearly suggest a single moving metal mechanism-presented in an accompanying (second) paper. A previously published 2.98 Å structure of a yeast topoisomerase II, which has static disorder around a crystallographic twofold axis, was published as containing two metals at one active site. Re-refined coordinates of this 2.98 Å yeast structure are consistent with other type IIA topoisomerase structures in only having one metal ion at each of the two different active sites.
- Medicines Discovery Institute, Cardiff University, Cardiff CF10 3AT, UK.
Organizational Affiliation: