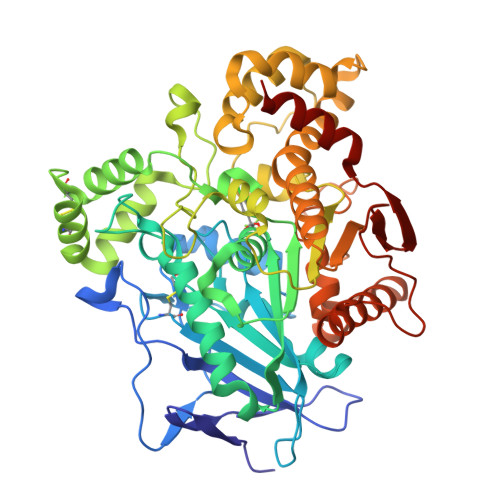
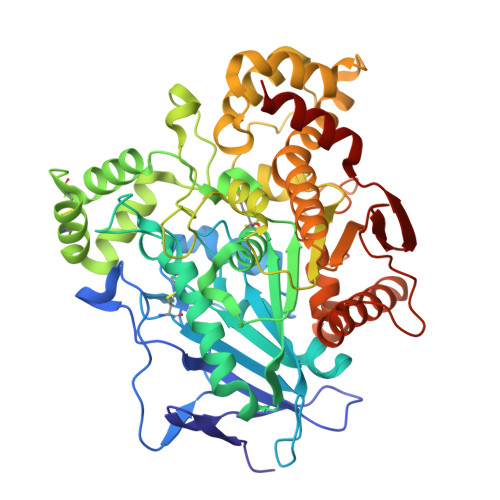
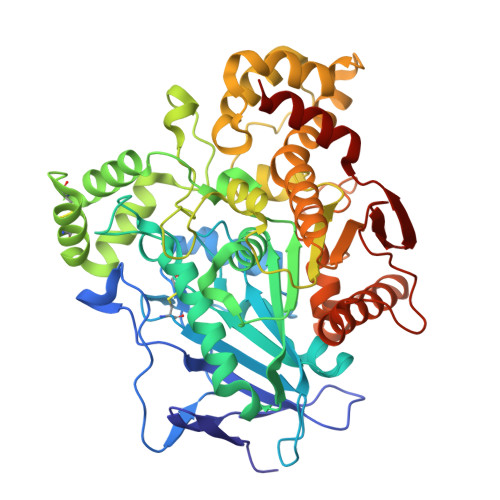
The study of halogen effect on the reactivity of the serine-targeting covalent warheads.
Gai, C., Zhang, Y., Zhang, S., Hu, X., Song, Y.Q., Zhuang, X., Chai, X., Zou, Y., Ge, G.B., Zhao, Q.(2024) Front Chem 12: 1504453-1504453
- PubMed: 39691824
- DOI: https://doi.org/10.3389/fchem.2024.1504453
- Primary Citation of Related Structures:
9KWL, 9KWM - PubMed Abstract:
Halogens favorably contributes to the drug potency and metabolic stability via electrostatic interactions. Herein, the halogen effects on the reactivity of the halogenated 2,2,2-trifluoroacetophenones as serine-targeting covalent warheads were investigated. Our results showed that introducing halogen atoms, especially Cl or Br, into the phenyl scaffold would influence the electron density around the ring, which led to different time-dependent inhibition response to the target serine hydrolase (hCES1A). Co-crystallography analysis not only verified that halogenated molecules preferred to form covalent adducts, but also provided the conformational information for the design of covalent inhibitors targeting to hCES1A protein for the treatment of drug-induced acute enteritis.
Organizational Affiliation:
Organic Chemistry Group, College of Pharmacy, Naval Medical University, Shanghai, China.