Alpha1/Beta Heteromeric Glycine receptor in the presence of 0.200 mM strychnine and 2000 nM ivermectin
Gibbs, E., Chakrapani, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycine receptor, alpha 1 | A, B [auth D], C, D [auth B] | 458 | Danio rerio | Mutation(s): 0 Gene Names: glra1 | 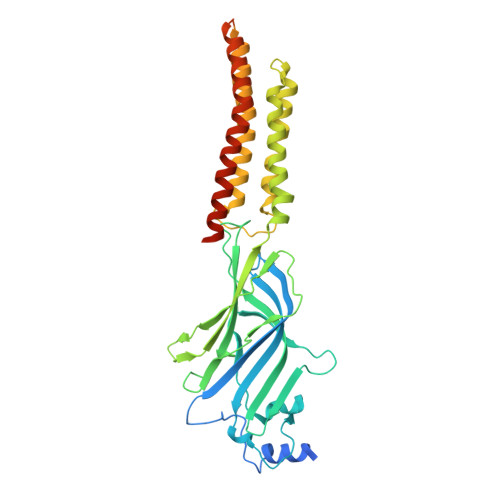 |
UniProt | |||||
Find proteins for B3DHZ5 (Danio rerio) Explore B3DHZ5 Go to UniProtKB: B3DHZ5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B3DHZ5 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycine receptor beta subunit 2 | 591 | Danio rerio | Mutation(s): 0 Gene Names: glrbb, glrb2 | 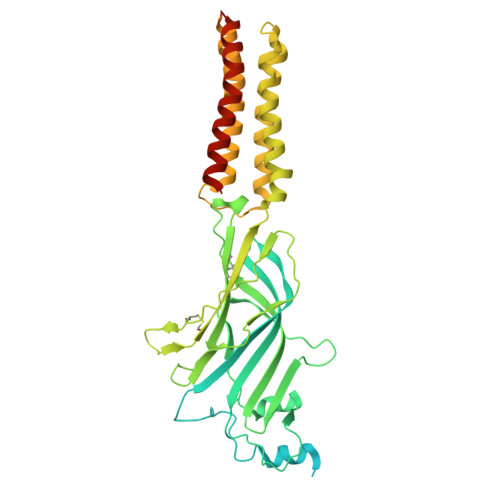 | |
UniProt | |||||
Find proteins for Q6DC22 (Danio rerio) Explore Q6DC22 Go to UniProtKB: Q6DC22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6DC22 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IVM (Subject of Investigation/LOI) Query on IVM | H [auth A], I [auth A], L [auth D], N [auth C], Q [auth B] | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17
-oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy
clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside C48 H74 O14 AZSNMRSAGSSBNP-XPNPUAGNSA-N |  | ||
| SY9 (Subject of Investigation/LOI) Query on SY9 | G [auth A], K [auth D], M [auth C], P [auth C], S [auth B] | STRYCHNINE C21 H22 N2 O2 QMGVPVSNSZLJIA-FVWCLLPLSA-N |  | ||
| NAG Query on NAG | F [auth A] J [auth D] O [auth C] R [auth B] T [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19.2_4158 |
| RECONSTRUCTION | RELION |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |