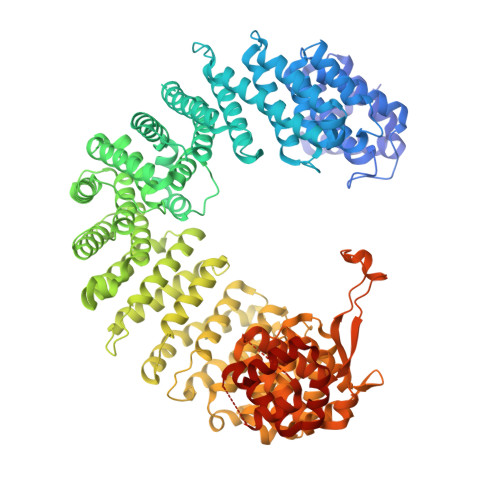
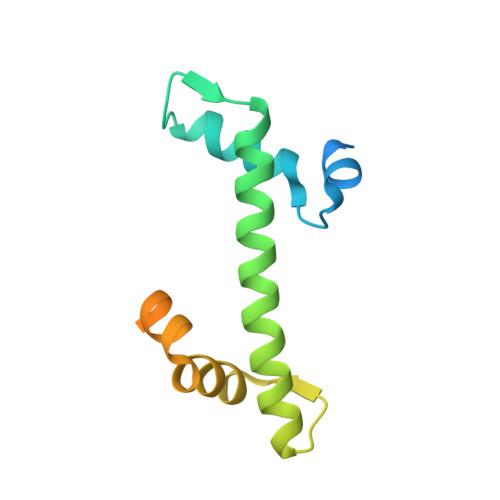
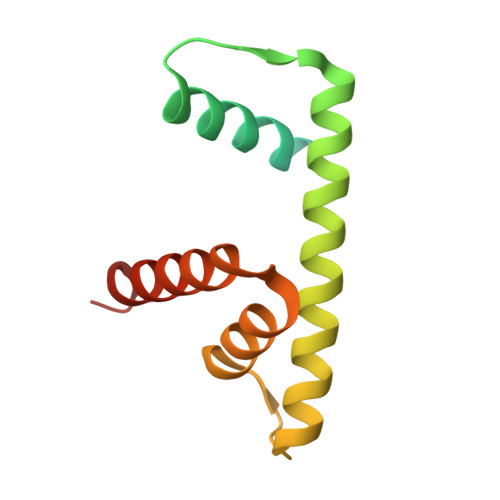
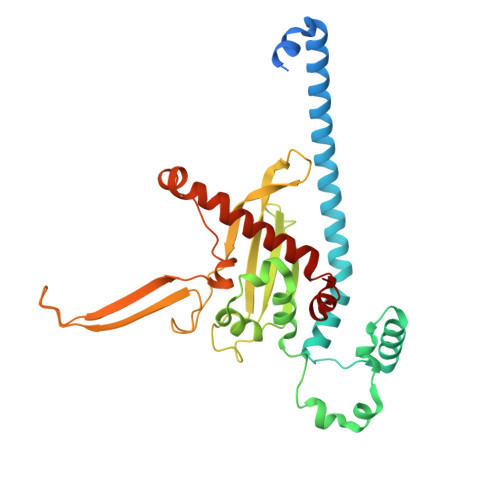
Nap1 and Kap114 co-chaperone H2A-H2B and facilitate targeted histone release in the nucleus.
Fung, H.Y.J., Jiou, J., Niesman, A.B., Bernardes, N.E., Chook, Y.M.(2025) J Cell Biol 224
- PubMed: 39601790
- DOI: https://doi.org/10.1083/jcb.202408193
- Primary Citation of Related Structures:
9B23, 9B31, 9B3F, 9B3I - PubMed Abstract:
Core histones, synthesized and processed in the cytoplasm, must be chaperoned as they are transported into the nucleus for nucleosome assembly. The importin Kap114 transports H2A-H2B into the yeast nucleus, where RanGTP facilitates histone release. Kap114 and H2A-H2B also bind the histone chaperone Nap1, but how Nap1 and Kap114 cooperate in transport and nucleosome assembly remains unclear. Here, biochemical and structural analyses show that Kap114, H2A-H2B, and a Nap1 dimer (Nap12) associate in the absence and presence of RanGTP to form equimolar complexes. A previous study had shown that RanGTP reduces Kap114's ability to chaperone H2A-H2B, but a new cryo-EM structure of the Nap12•H2A-H2B•Kap114•RanGTP complex explains how both Kap114 and Nap12 interact with H2A-H2B, restoring its chaperoning within the assembly while effectively depositing it into nucleosomes. Together, our results suggest that Kap114 and Nap12 provide a sheltered path that facilitates the transfer of H2A-H2B from Kap114 to Nap12, ultimately directing its specific deposition into nucleosomes.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: