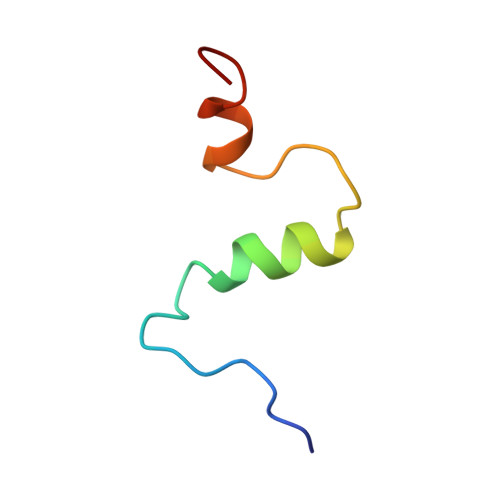
Structure of amyloid A4-(1-40)-peptide of Alzheimer's disease.
Sticht, H., Bayer, P., Willbold, D., Dames, S., Hilbich, C., Beyreuther, K., Frank, R.W., Rosch, P.(1995) Eur J Biochem 233: 293-298
- PubMed: 7588758
- DOI: https://doi.org/10.1111/j.1432-1033.1995.293_1.x
- Primary Citation of Related Structures:
1AML - PubMed Abstract:
One of the principle peptide components of the amyloid plaque deposits of Alzheimer's disease in humans is the 40-amino-acid peptide beta-amyloid A4-(1-40)-peptide. The full-length A4-(1-40)-peptide was chemically synthesized and the solution structure determined by two-dimensional nuclear magnetic resonance spectroscopy and restrained molecular-dynamics calculations. Synthetic human A4-(1-40)-peptide was soluble and non-aggregating for several days in 40% (by vol.) trifluoroethanol/water. All spin systems could be unambiguously assigned, and a total of 203 sequential and medium-range cross-peaks were found in the NOESY (nuclear Overhauser enhancement spectroscopy) spectrum. Long-range NOE cross-peaks that would indicate tertiary structure of the peptide were absent. The main secondary-structure elements found by chemical-shift analysis, sequential and medium-range NOESY data, and NOE-based restrained molecular-dynamics calculations were two helices, Gln15-Asp23 and Ile31-Met35, whereas the rest of the peptide was in random-coil conformation. A similar secondary structure is suggested for the aggregation part of prions, the postulated causative agents of the transmissible spongiform encephalopathy. The sequence of the helical part of prion proteins was observed to be remarkably similar to the sequence of the helical part of human A4-(1-40)-peptide.
- Lehrstuhl für Biopolymere, Universität Bayreuth, Bayreuth, Germany.
Organizational Affiliation: