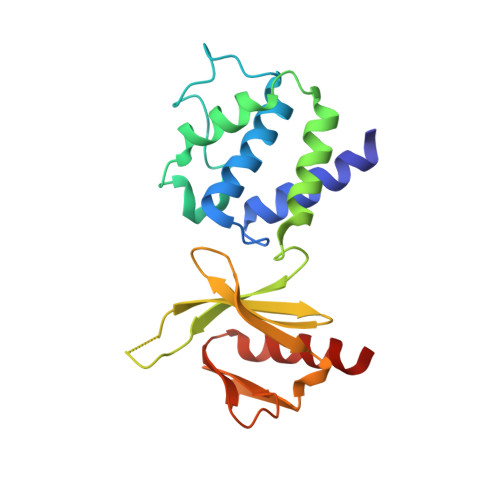
Structural Determinants of Integrin Recognition by Talin
Garcia-Alvarez, B., de Pereda, J.M., Calderwood, D.A., Ulmer, T.S., Critchley, D., Campbell, I.D., Ginsberg, M.H., Liddington, R.C.(2003) Mol Cell 11: 49-58
- PubMed: 12535520
- DOI: https://doi.org/10.1016/s1097-2765(02)00823-7
- Primary Citation of Related Structures:
1MIX, 1MIZ, 1MK7, 1MK9 - PubMed Abstract:
The binding of cytoplasmic proteins, such as talin, to the cytoplasmic domains of integrin adhesion receptors mediates bidirectional signal transduction. Here we report the crystal structure of the principal integrin binding and activating fragment of talin, alone and in complex with fragments of the beta 3 integrin tail. The FERM (four point one, ezrin, radixin, and moesin) domain of talin engages integrins via a novel variant of the canonical phosphotyrosine binding (PTB) domain-NPxY ligand interaction that may be a prototype for FERM domain recognition of transmembrane receptors. In combination with NMR and mutational analysis, our studies reveal the critical interacting elements of both talin and the integrin beta 3 tail, providing structural paradigms for integrin linkage to the cell interior.
- Program on Cell Adhesion, The Burnham Institute, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: