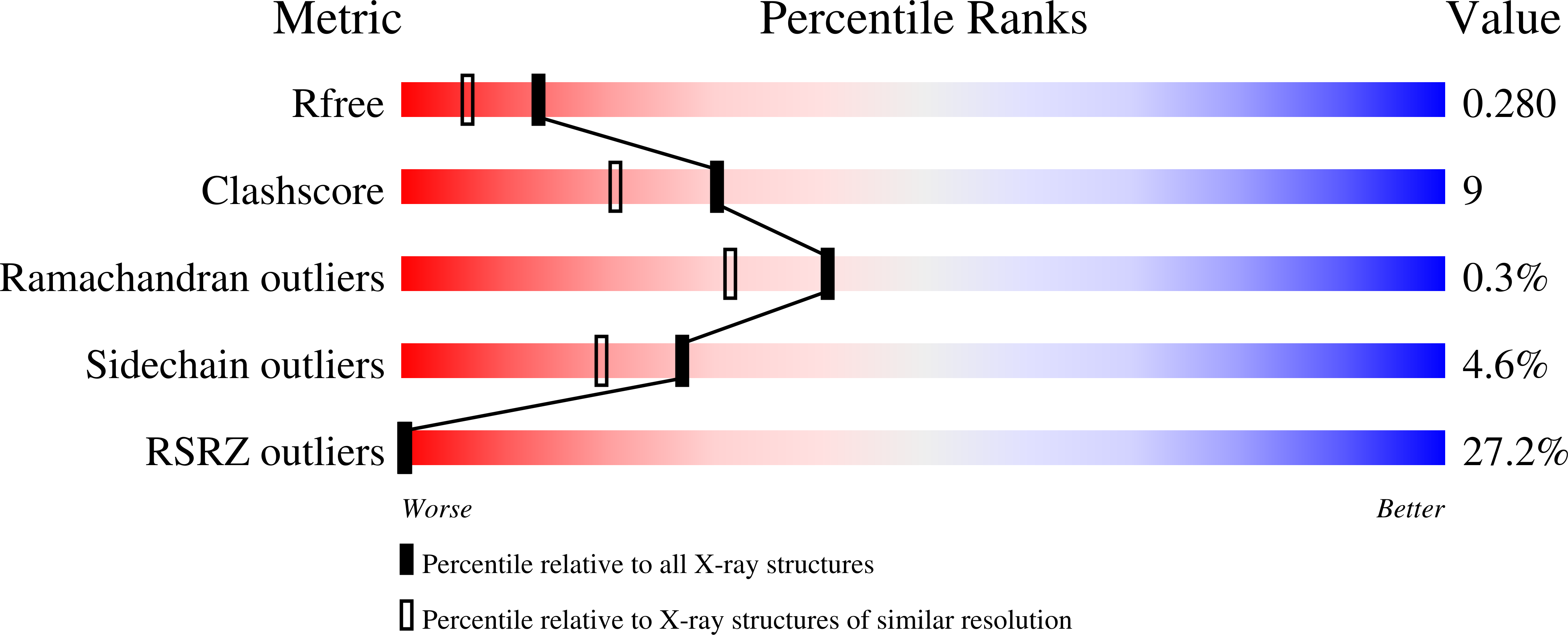
Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Couture, J.F., Collazo, E., Ortiz-Tello, P.A., Brunzelle, J.S., Trievel, R.C.(2007) Nat Struct Mol Biol 14: 689-695
- PubMed: 17589523
- DOI: https://doi.org/10.1038/nsmb1273
- PubMed Abstract:
JMJD2A is a JmjC histone demethylase (HDM) that catalyzes the demethylation of di- and trimethylated Lys9 and Lys36 in histone H3 (H3K9me2/3 and H3K36me2/3). Here we present the crystal structures of the JMJD2A catalytic domain in complex with H3K9me3, H3K36me2 and H3K36me3 peptides. The structures reveal that histone substrates are recognized through a network of backbone hydrogen bonds and hydrophobic interactions that deposit the trimethyllysine into the active site. The trimethylated epsilon-ammonium cation is coordinated within a methylammonium-binding pocket through carbon-oxygen (CH...O) hydrogen bonds that position one of the zeta-methyl groups adjacent to the Fe(II) center for hydroxylation and demethylation. Mutations of the residues comprising this pocket abrogate demethylation by JMJD2A, with the exception of an S288A substitution, which augments activity, particularly toward H3K9me2. We propose that this residue modulates the methylation-state specificities of JMJD2 enzymes and other trimethyllysine-specific JmjC HDMs.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, 1150 West Medical Center Drive, Ann Arbor, Michigan 48109-0606, USA.