Structural insight into the membrane insertion of tail-anchored proteins by Get3
Yamagata, A., Mimura, H., Sato, Y., Yamashita, M., Yoshikawa, A., Fukai, S.(2010) Genes Cells 15: 29-41
- PubMed: 20015340
- DOI: https://doi.org/10.1111/j.1365-2443.2009.01362.x
- Primary Citation of Related Structures:
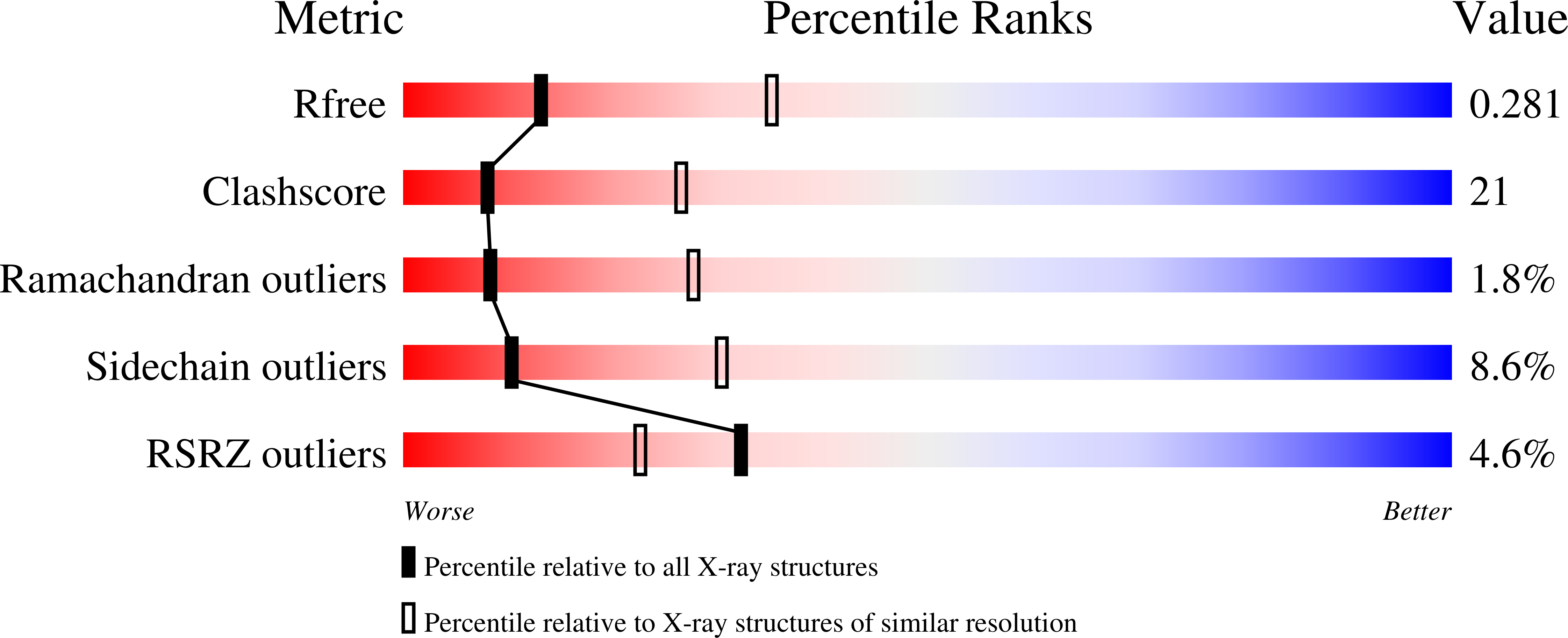
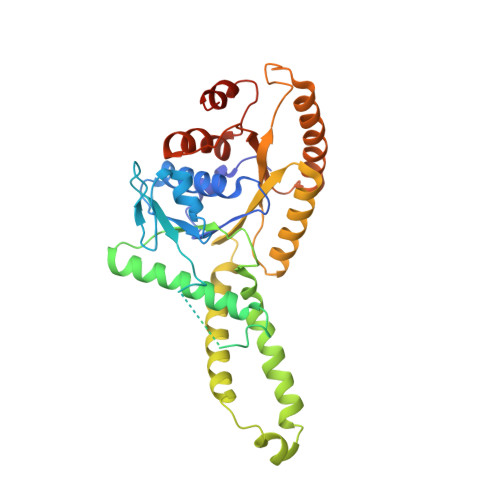
3A36, 3A37 - PubMed Abstract:
Tail anchored (TA) proteins, which are important for numerous cellular processes, are defined by a single transmembrane domain (TMD) near the C-terminus. The membrane insertion of TA proteins is mediated by the highly conserved ATPase Get3. Here we report the crystal structures of Get3 in ADP-bound and nucleotide-free forms at 3.0 A and 2.8 A resolutions, respectively. Get3 consists of a nucleotide binding domain and a helical domain. Both structures exhibit a Zn(2+)-mediated homodimer in a head-to-head orientation, representing an open dimer conformation. Our cross-link experiments indicated the closed dimer-stimulating ATP hydrolysis, which might be coupled with TA-protein release. Further, our coexpression-based binding assays using a model TA protein Sec22p revealed the direct interaction between the helical domain of Get3 and the Sec22p TMD. This interaction is independent of ATP and dimer formation. Finally, we propose a structural mechanism that links ATP hydrolysis with the TA-protein insertion mediated by the conserved DTAPTGH motif.
Organizational Affiliation:
Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo 113-0032, Japan.