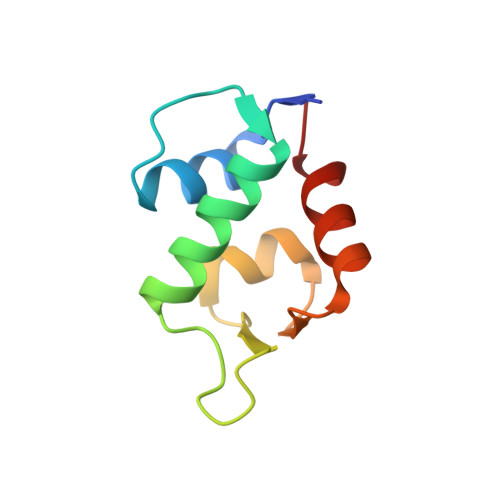
Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX.
Phan, J., Li, Z., Kasprzak, A., Li, B., Sebti, S., Guida, W., Schonbrunn, E., Chen, J.(2010) J Biological Chem 285: 2174-2183
- PubMed: 19910468
- DOI: https://doi.org/10.1074/jbc.M109.073056
- Primary Citation of Related Structures:
3JZO, 3JZP, 3JZQ, 3JZR, 3JZS - PubMed Abstract:
MDM2 and MDMX function as key regulators of p53 by binding to its N terminus, inhibiting its transcriptional activity, and promoting degradation. MDM2 and MDMX overexpression or hyperactivation directly contributes to the loss of p53 function during the development of nearly 50% of human cancers. Recent studies showed that disrupting p53-MDM2 and p53-MDMX interactions can lead to robust activation of p53 but also revealed a need to develop novel dual specific or MDMX-specific inhibitors. Using phage display we identified a 12-residue peptide (pDI) with inhibitory activity against MDM2 and MDMX. The co-crystal structures of the pDI and a single mutant derivative (pDI6W) liganded with the N-terminal domains of human MDMX and MDM2 served as the basis for the design of 11 distinct pDI-derivative peptides that were tested for inhibitory potential. The best derivative (termed pDIQ) contained four amino acid substitutions and exhibited a 5-fold increase in potency over the parent peptide against both MDM2 (IC(50) = 8 nm) and MDMX (IC(50) = 110 nm). Further structural studies revealed key molecular features enabling the high affinity binding of the pDIQ to these proteins. These include large conformational changes of the pDIQ to reach into a hydrophobic site unique to MDMX. The findings suggest new strategies toward the rational design of small molecule inhibitors efficiently targeting MDMX.
- Molecular Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, USA.
Organizational Affiliation: