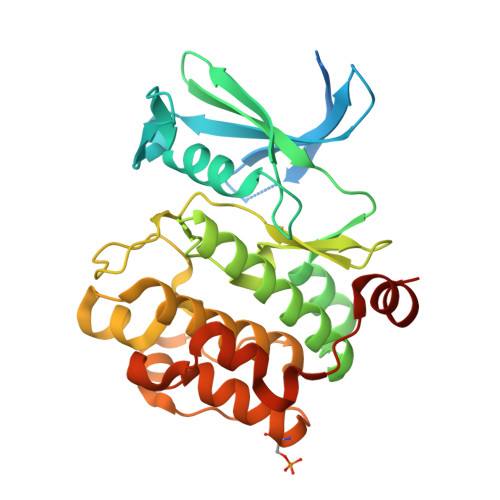
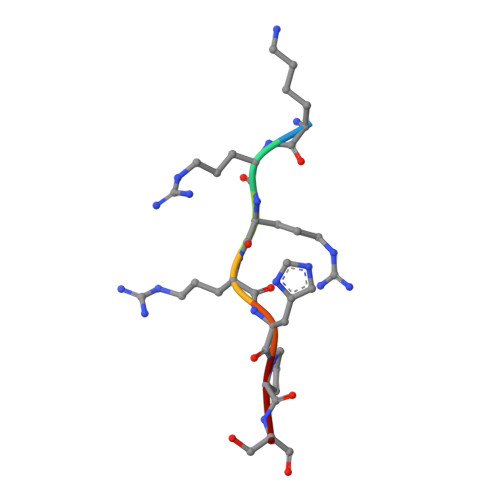
New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Lopez-Ramos, M., Prudent, R., Moucadel, V., Sautel, C.F., Barette, C., Lafanechere, L., Mouawad, L., Grierson, D., Schmidt, F., Florent, J.C., Filippakopoulos, P., Bullock, A.N., Knapp, S., Reiser, J.B., Cochet, C.(2010) FASEB J 24: 3171-3185
- PubMed: 20400536
- DOI: https://doi.org/10.1096/fj.09-143743
- Primary Citation of Related Structures:
3MA3, 3MB6, 3MB7 - PubMed Abstract:
Protein kinase casein kinase 2 (CK2) is a serine/threonine kinase with evidence of implication in growth dysregulation and apoptosis resistance, making it a relevant target for cancer therapy. Several CK2 inhibitors have been developed showing variable efficiency, emphasizing the need to expand the chemical diversity of those inhibitors. We report the identification and characterization of 2,8-difurandicarboxylic acid derivatives as a new class of nanomolar ATP-competitive inhibitors. Selectivity profiling pointed out proviral insertion Moloney virus kinases (Pim kinases) as the only other kinases that are significantly inhibited. By combining structure-activity relationship analysis with structural determination, we were able to determine the binding mode of these inhibitors for both kinases and to explain their strong inhibitory potency. Essential chemical features necessary for activity on both kinases were then identified. The described compounds are not cell permeable: however, they could provide a lead for developing novel inhibitors usable also in vivo. Given the similar but not redundant pathophysiological functions of CK2 and Pim family members, such inhibitors would provide new attractive leads for targeted cancer therapy. This work highlights that 2 functionally related kinases from different kinome branches display exquisite sensitivity to a common inhibitor.
- Centre de Recherche, Institut Curie, Paris, France.
Organizational Affiliation: