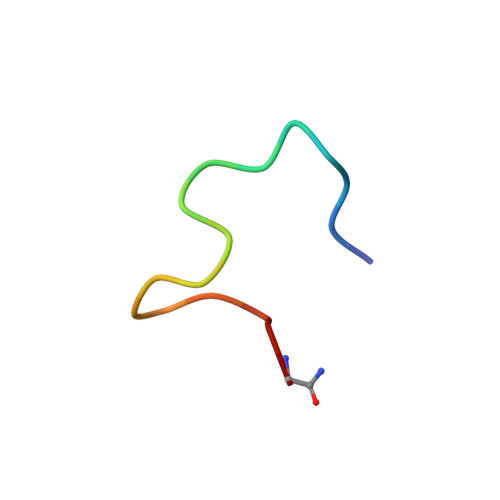
Improving binding affinity and stability of Peptide ligands by substituting glycines with d-amino acids.
Chen, S., Gfeller, D., Buth, S.A., Michielin, O., Leiman, P.G., Heinis, C.(2013) Chembiochem 14: 1316-1322
- PubMed: 23828687
- DOI: https://doi.org/10.1002/cbic.201300228
- Primary Citation of Related Structures:
4JK5, 4JK6 - PubMed Abstract:
Improving the binding affinity and/or stability of peptide ligands often requires testing of large numbers of variants to identify beneficial mutations. Herein we propose a type of mutation that promises a high success rate. In a bicyclic peptide inhibitor of the cancer-related protease urokinase-type plasminogen activator (uPA), we observed a glycine residue that has a positive ϕ dihedral angle when bound to the target. We hypothesized that replacing it with a D-amino acid, which favors positive ϕ angles, could enhance the binding affinity and/or proteolytic resistance. Mutation of this specific glycine to D-serine in the bicyclic peptide indeed improved inhibitory activity (1.75-fold) and stability (fourfold). X-ray-structure analysis of the inhibitors in complex with uPA showed that the peptide backbone conformation was conserved. Analysis of known cyclic peptide ligands showed that glycine is one of the most frequent amino acids, and that glycines with positive ϕ angles are found in many protein-bound peptides. These results suggest that the glycine-to-D-amino acid mutagenesis strategy could be broadly applied.
- Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne, BCH 5305 (Batochime), Avenue Forel 2, 1015 Lausanne, Switzerland.
Organizational Affiliation: