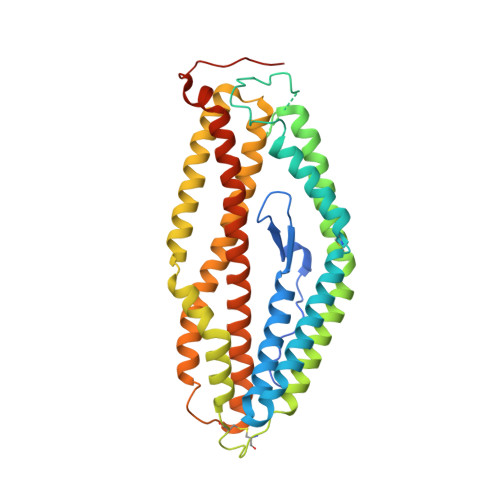
Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes.
Chen, L., Xu, Y., Healer, J., Thompson, J.K., Smith, B.J., Lawrence, M.C., Cowman, A.F.(2014) Elife 3
- PubMed: 25296023
- DOI: https://doi.org/10.7554/eLife.04187
- Primary Citation of Related Structures:
4WAT - PubMed Abstract:
Plasmodium falciparum causes the most severe form of malaria in humans and is responsible for over 700,000 deaths annually. It is an obligate intracellular parasite and invades erythrocytes where it grows in a relatively protected niche. Invasion of erythrocytes is essential for parasite survival and this involves interplay of multiple protein–protein interactions. One of the most important interactions is binding of parasite invasion ligand families EBLs and PfRhs to host receptors on the surface of erythrocytes. PfRh5 is the only essential invasion ligand within the PfRh family and is an important vaccine candidate. PfRh5 binds the host receptor basigin. In this study, we have determined the crystal structure of PfRh5 using diffraction data to 2.18 Å resolution. PfRh5 exhibits a novel fold, comprising nine mostly anti-parallel α-helices encasing an N-terminal β-hairpin, with the overall shape being an elliptical disk. This is the first three-dimensional structure determined for the PfRh family of proteins. DOI: http://dx.doi.org/10.7554/eLife.04187.001
- Division of Infection and Immunity, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia.
Organizational Affiliation: