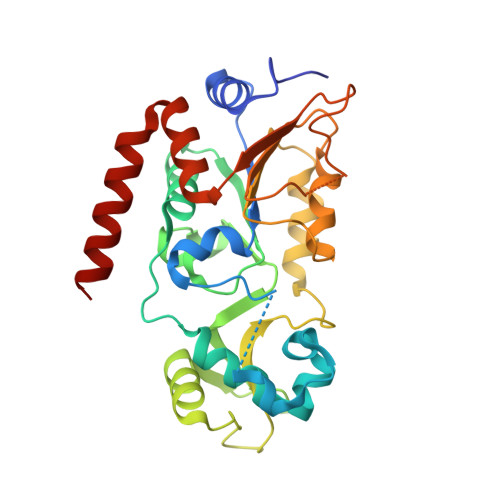
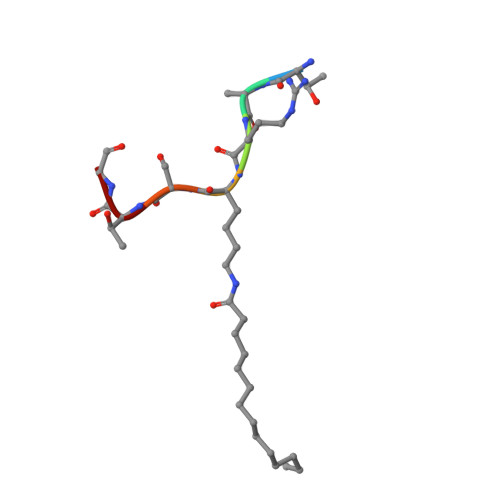
Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Feldman, J.L., Dittenhafer-Reed, K.E., Kudo, N., Thelen, J.N., Ito, A., Yoshida, M., Denu, J.M.(2015) Biochemistry 54: 3037-3050
- PubMed: 25897714
- DOI: https://doi.org/10.1021/acs.biochem.5b00150
- Primary Citation of Related Structures:
4Y6L, 4Y6O, 4Y6Q - PubMed Abstract:
Acylation of lysine is an important protein modification regulating diverse biological processes. It was recently demonstrated that members of the human Sirtuin family are capable of catalyzing long chain deacylation, in addition to the well-known NAD(+)-dependent deacetylation activity [Feldman, J. L., Baeza, J., and Denu, J. M. (2013) J. Biol. Chem. 288, 31350-31356]. Here we provide a detailed kinetic and structural analysis that describes the interdependence of NAD(+)-binding and acyl-group selectivity for a diverse series of human Sirtuins, SIRT1-SIRT3 and SIRT6. Steady-state and rapid-quench kinetic analyses indicated that differences in NAD(+) saturation and susceptibility to nicotinamide inhibition reflect unique kinetic behavior displayed by each Sirtuin and depend on acyl substrate chain length. Though the rate of nucleophilic attack of the 2'-hydroxyl on the C1'-O-alkylimidate intermediate varies with acyl substrate chain length, this step remains rate-determining for SIRT2 and SIRT3; however, for SIRT6, this step is no longer rate-limiting for long chain substrates. Cocrystallization of SIRT2 with myristoylated peptide and NAD(+) yielded a co-complex structure with reaction product 2'-O-myristoyl-ADP-ribose, revealing a latent hydrophobic cavity to accommodate the long chain acyl group, and suggesting a general mechanism for long chain deacylation. Comparing two separately determined co-complex structures containing either a myristoylated peptide or 2'-O-myristoyl-ADP-ribose indicates there are conformational changes at the myristoyl-ribose linkage with minimal structural differences in the enzyme active site. During the deacylation reaction, the fatty acyl group is held in a relatively fixed position. We describe a kinetic and structural model to explain how various Sirtuins display unique acyl substrate preferences and how different reaction kinetics influence NAD(+) dependence. The biological implications are discussed.
- Department of Biomolecular Chemistry, University of Wisconsin-Madison, Madison, Wisconsin 53715.
Organizational Affiliation: