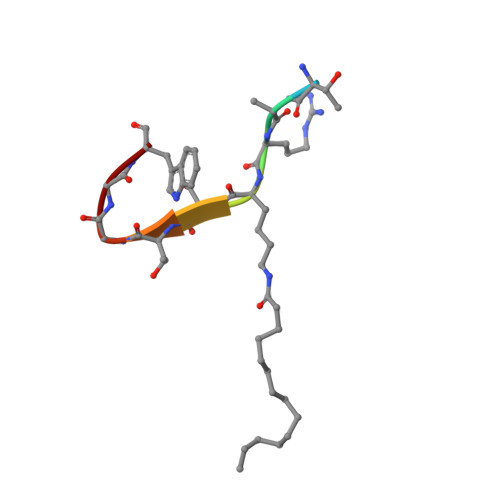
Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Gai, W., Li, H., Jiang, H., Long, Y., Liu, D.(2016) FEBS Lett 590: 3019-3028
- PubMed: 27501476
- DOI: https://doi.org/10.1002/1873-3468.12345
- Primary Citation of Related Structures:
5BWN, 5BWO - PubMed Abstract:
SIRT1-7 play important roles in many biological processes and age-related diseases. In addition to a NAD(+) -dependent deacetylase activity, they can catalyze several other reactions, including the hydrolysis of long-chain fatty acyl lysine. To study the binding modes of sirtuins to long-chain acyl lysines, we solved the crystal structures of SIRT3 bound to either a H3K9-myristoylated- or a H3K9-palmitoylated peptide. Interaction of SIRT3 with the palmitoyl group led to unfolding of the α3-helix. The myristoyl and palmitoyl groups bind to the C-pocket and an allosteric site near the α3-helix, respectively. We found that the residues preceding the α3-helix determine the size of the C-pocket. The flexibility of the α2-α3 loop and the plasticity of the α3-helix affect the interaction with long-chain acyl lysine.
- Department of Pharmacology III, Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, China.
Organizational Affiliation: