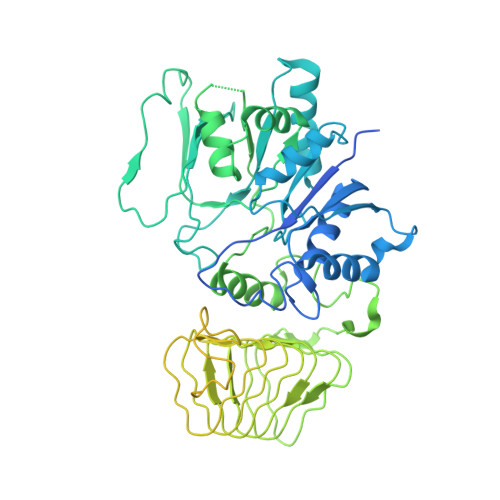
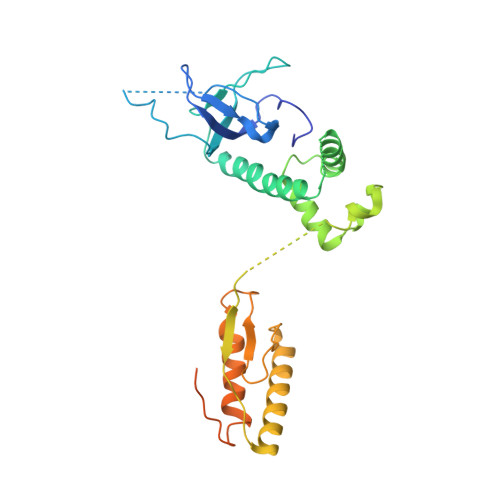
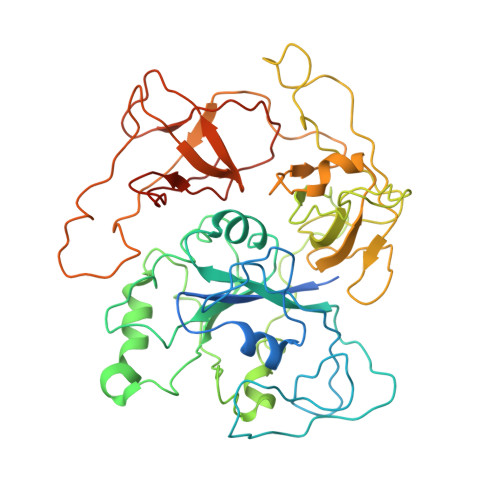
Structural basis for eIF2B inhibition in integrated stress response.
Kashiwagi, K., Yokoyama, T., Nishimoto, M., Takahashi, M., Sakamoto, A., Yonemochi, M., Shirouzu, M., Ito, T.(2019) Science 364: 495-499
- PubMed: 31048492
- DOI: https://doi.org/10.1126/science.aaw4104
- Primary Citation of Related Structures:
6JLY, 6JLZ, 6K71, 6K72 - PubMed Abstract:
A core event in the integrated stress response, an adaptive pathway common to all eukaryotic cells in response to various stress stimuli, is the phosphorylation of eukaryotic translation initiation factor 2 (eIF2). Normally, unphosphorylated eIF2 transfers the methionylated initiator tRNA to the ribosome in a guanosine 5'-triphosphate-dependent manner. By contrast, phosphorylated eIF2 inhibits its specific guanine nucleotide exchange factor, eIF2B. To elucidate how the eIF2 phosphorylation status regulates the eIF2B activity, we determined cryo-electron microscopic and crystallographic structures of eIF2B in complex with unphosphorylated or phosphorylated eIF2. The unphosphorylated and phosphorylated forms of eIF2 bind to eIF2B in completely different manners: the nucleotide exchange-active and -inactive modes, respectively. These structures explain how phosphorylated eIF2 dominantly inhibits the nucleotide exchange activity of eIF2B.
- RIKEN Center for Biosystems Dynamics Research, Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: