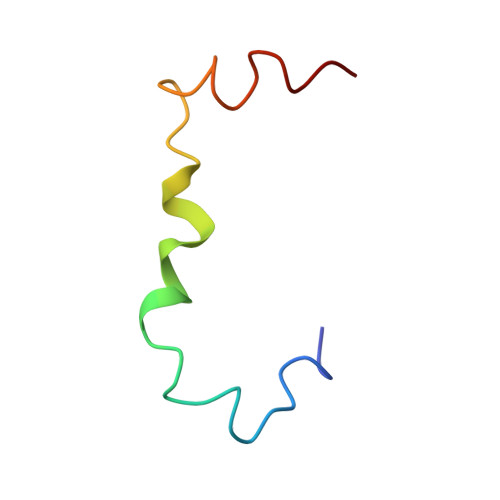
Exploring the Early Stages of the Amyloid A beta (1-42) Peptide Aggregation Process: An NMR Study.
Santoro, A., Grimaldi, M., Buonocore, M., Stillitano, I., D'Ursi, A.M.(2021) Pharmaceuticals (Basel) 14
- PubMed: 34451828
- DOI: https://doi.org/10.3390/ph14080732
- Primary Citation of Related Structures:
6SZF - PubMed Abstract:
Alzheimer's disease (AD) is a neurodegenerative pathology characterized by the presence of neurofibrillary tangles and amyloid plaques, the latter mainly composed of Aβ(1-40) and Aβ(1-42) peptides. The control of the Aβ aggregation process as a therapeutic strategy for AD has prompted the interest to investigate the conformation of the Aβ peptides, taking advantage of computational and experimental techniques. Mixtures composed of systematically different proportions of HFIP and water have been used to monitor, by NMR, the conformational transition of the Aβ(1-42) from soluble α-helical structure to β-sheet aggregates. In the previous studies, 50/50 HFIP/water proportion emerged as the solution condition where the first evident Aβ(1-42) conformational changes occur. In the hypothesis that this solvent reproduces the best condition to catch transitional helical-β-sheet Aβ(1-42) conformations, in this study, we report an extensive NMR conformational analysis of Aβ(1-42) in 50/50 HFIP/water v / v . Aβ(1-42) structure was solved by us, giving evidence that the evolution of Aβ(1-42) peptide from helical to the β-sheet may follow unexpected routes. Molecular dynamics simulations confirm that the structural model we calculated represents a starting condition for amyloid fibrils formation.
- Department of Pharmacy, University of Salerno, Via Giovanni Paolo II, 132-84084 Fisciano, SA, Italy.
Organizational Affiliation: