Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
Tassone, G., Di Pisa, F., Benvenuti, M., De Luca, F., Pozzi, C., Docquier, J.D., Mangani, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase | 265 | Klebsiella pneumoniae | Mutation(s): 0 Gene Names: bla OXA-48, blaOXA-48, KPE71T_00045 EC: 3.5.2.6 | 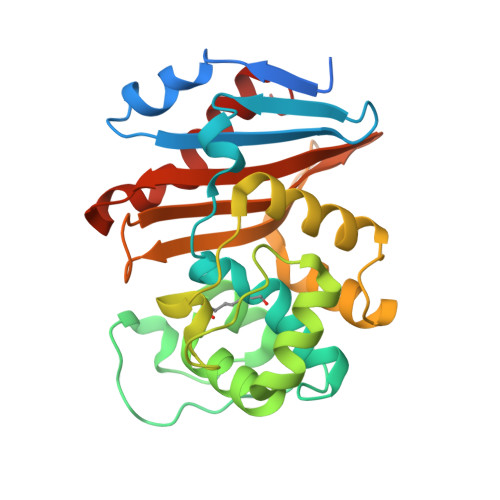 | |
UniProt | |||||
Find proteins for Q6XEC0 (Klebsiella pneumoniae) Explore Q6XEC0 Go to UniProtKB: Q6XEC0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6XEC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase | 265 | Klebsiella pneumoniae | Mutation(s): 0 Gene Names: bla OXA-48, bla_1, bla_2, bla_5, blaOXA-48, B6R99_29845, GJD56_28020, GJJ04_29145, KPE71T_00045, SAMEA3538918_02768... EC: 3.5.2.6 | 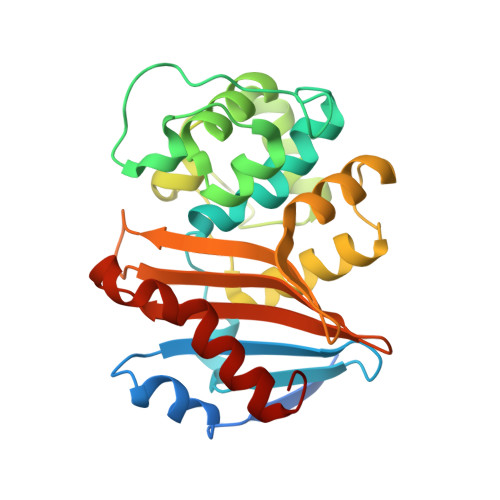 | |
UniProt | |||||
Find proteins for Q6XEC0 (Klebsiella pneumoniae) Explore Q6XEC0 Go to UniProtKB: Q6XEC0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6XEC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 2RG (Subject of Investigation/LOI) Query on 2RG | AA [auth F] CA [auth G] EA [auth H] I [auth A] M [auth B] | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid C22 H27 N3 O7 S RALHTEQYPKBGFO-SAGZMZRGSA-N |  | ||
| PGE Query on PGE | W [auth D] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | BA [auth F] DA [auth G] FA [auth H] GA [auth H] J [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | L [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| KCX Query on KCX | A, C, D, E, F A, C, D, E, F, G, H | L-PEPTIDE LINKING | C7 H14 N2 O4 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 63.834 | α = 90 |
| b = 162.839 | β = 90.59 |
| c = 108.158 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |