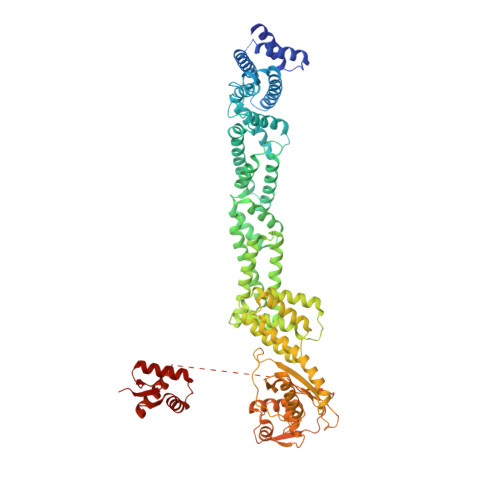
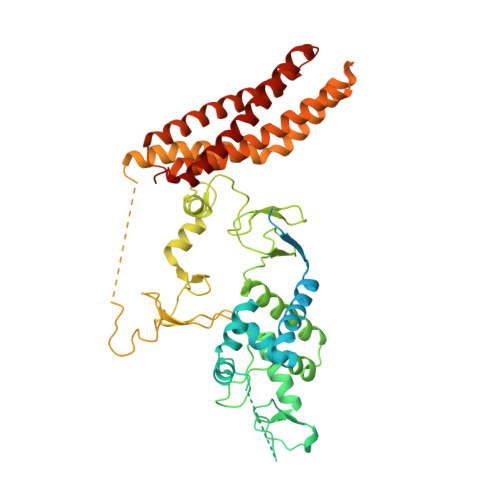
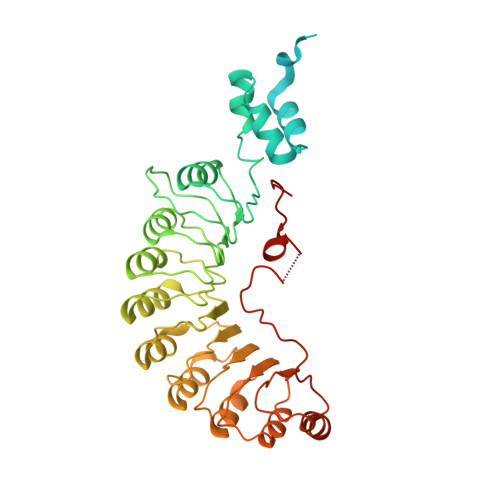
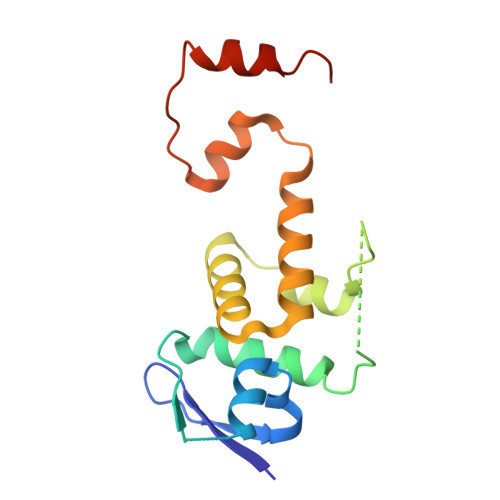
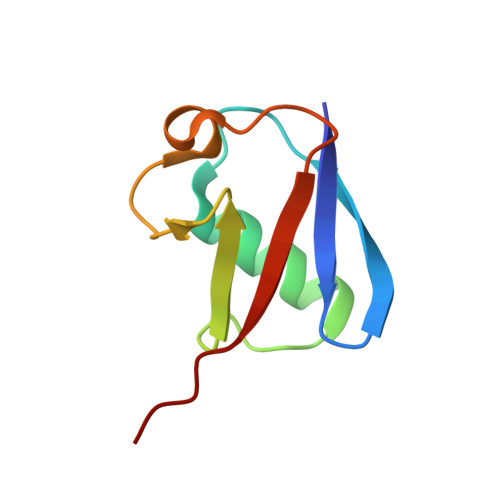
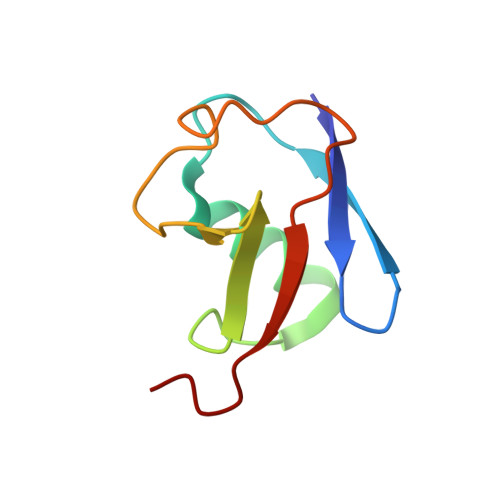
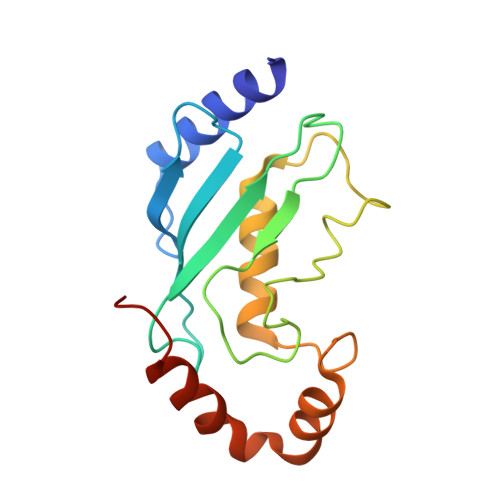
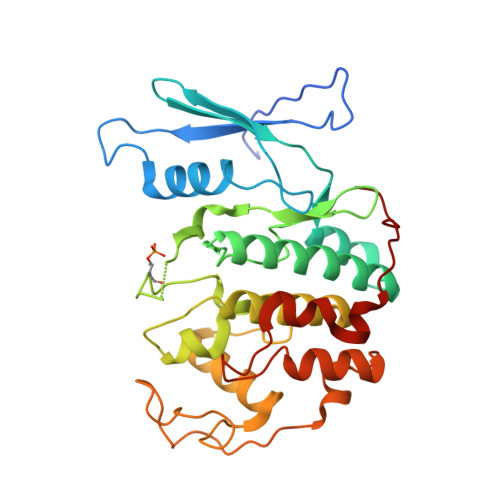
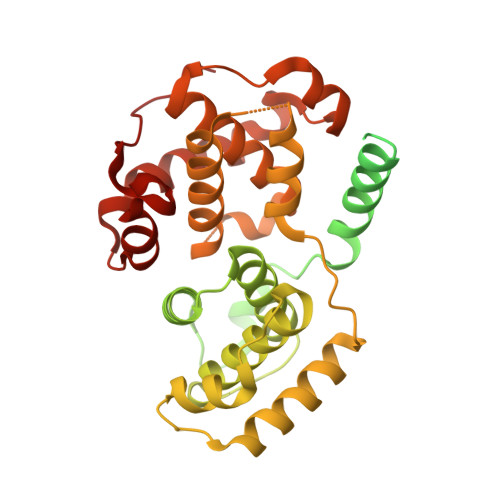
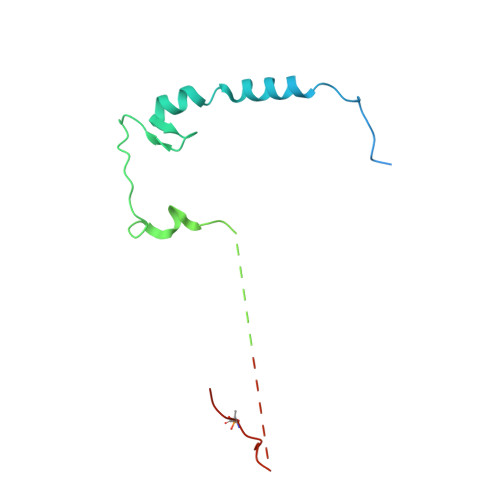
Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Horn-Ghetko, D., Krist, D.T., Prabu, J.R., Baek, K., Mulder, M.P.C., Klugel, M., Scott, D.C., Ovaa, H., Kleiger, G., Schulman, B.A.(2021) Nature 590: 671-676
- PubMed: 33536622
- DOI: https://doi.org/10.1038/s41586-021-03197-9
- Primary Citation of Related Structures:
7B5L, 7B5M, 7B5N, 7B5R, 7B5S - PubMed Abstract:
E3 ligases are typically classified by hallmark domains such as RING and RBR, which are thought to specify unique catalytic mechanisms of ubiquitin transfer to recruited substrates 1,2 . However, rather than functioning individually, many neddylated cullin-RING E3 ligases (CRLs) and RBR-type E3 ligases in the ARIH family-which together account for nearly half of all ubiquitin ligases in humans-form E3-E3 super-assemblies 3-7 . Here, by studying CRLs in the SKP1-CUL1-F-box (SCF) family, we show how neddylated SCF ligases and ARIH1 (an RBR-type E3 ligase) co-evolved to ubiquitylate diverse substrates presented on various F-box proteins. We developed activity-based chemical probes that enabled cryo-electron microscopy visualization of steps in E3-E3 ubiquitylation, initiating with ubiquitin linked to the E2 enzyme UBE2L3, then transferred to the catalytic cysteine of ARIH1, and culminating in ubiquitin linkage to a substrate bound to the SCF E3 ligase. The E3-E3 mechanism places the ubiquitin-linked active site of ARIH1 adjacent to substrates bound to F-box proteins (for example, substrates with folded structures or limited length) that are incompatible with previously described conventional RING E3-only mechanisms. The versatile E3-E3 super-assembly may therefore underlie widespread ubiquitylation.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: