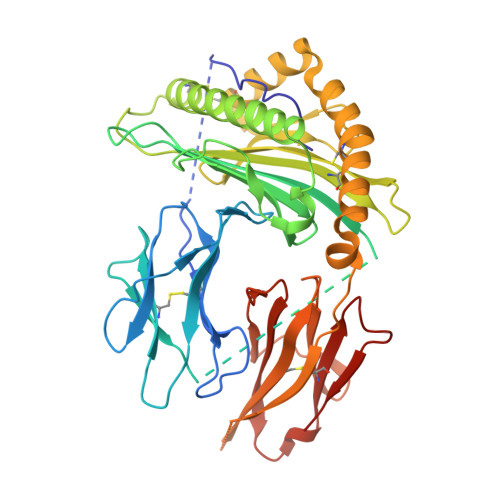
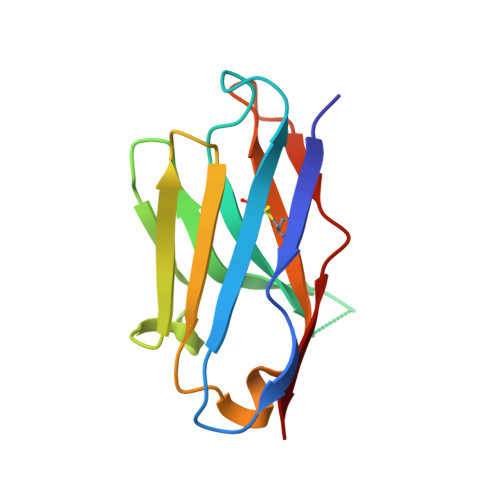
Effects of HLA single chain trimer design on peptide presentation and stability.
Finton, K.A.K., Rupert, P.B., Friend, D.J., Dinca, A., Lovelace, E.S., Buerger, M., Rusnac, D.V., Foote-McNabb, U., Chour, W., Heath, J.R., Campbell, J.S., Pierce, R.H., Strong, R.K.(2023) Front Immunol 14: 1170462-1170462
- PubMed: 37207206
- DOI: https://doi.org/10.3389/fimmu.2023.1170462
- Primary Citation of Related Structures:
7SQP, 7SR0, 7SR3, 7SR4, 7SR5, 7SRK, 7SSH, 7ST3, 7STG - PubMed Abstract:
MHC class I " single-chain trimer " molecules, coupling MHC heavy chain, β 2 -microglobulin, and a specific peptide into a single polypeptide chain, are widely used in research. To more fully understand caveats associated with this design that may affect its use for basic and translational studies, we evaluated a set of engineered single-chain trimers with combinations of stabilizing mutations across eight different classical and non-classical human class I alleles with 44 different peptides, including a novel human/murine chimeric design. While, overall, single-chain trimers accurately recapitulate native molecules, care was needed in selecting designs for studying peptides longer or shorter than 9-mers, as single-chain trimer design could affect peptide conformation. In the process, we observed that predictions of peptide binding were often discordant with experiment and that yields and stabilities varied widely with construct design. We also developed novel reagents to improve the crystallizability of these proteins and confirmed novel modes of peptide presentation.
- Division of Basic Science, Fred Hutchinson Cancer Research Center (FHCC), Seattle, WA, United States.
Organizational Affiliation: