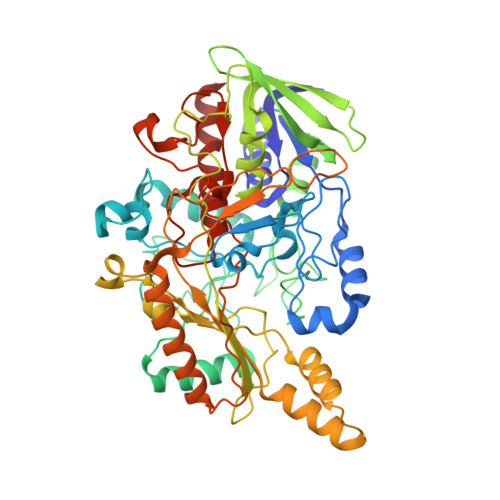
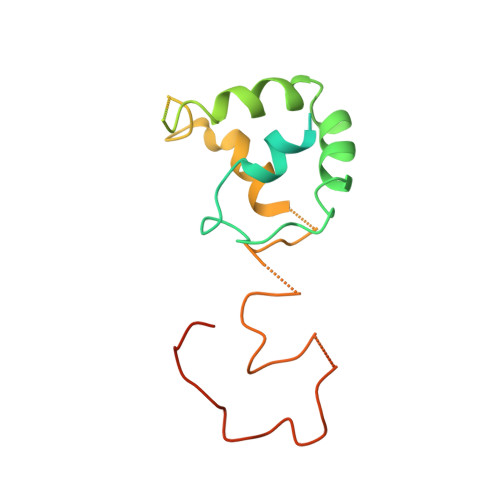
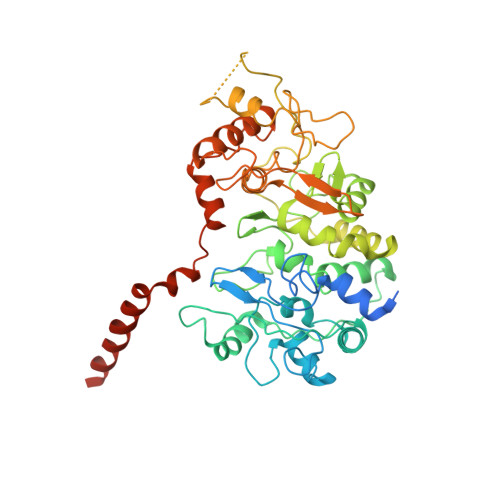
Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Suzuki, Y., Makino, F., Miyata, T., Tanaka, H., Namba, K., Kano, K., Sowa, K., Kitazumi, Y., Shirai, O.(2023) ACS Catal 13: 13828-13837