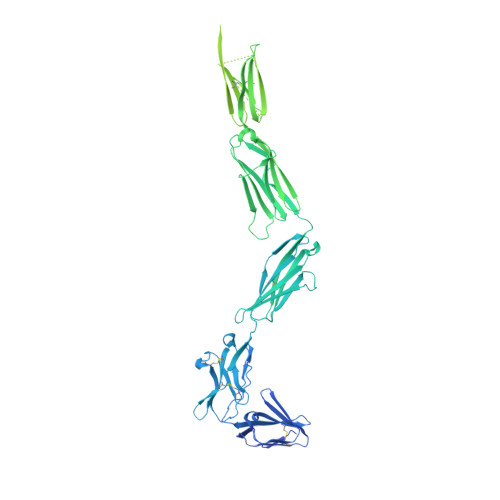
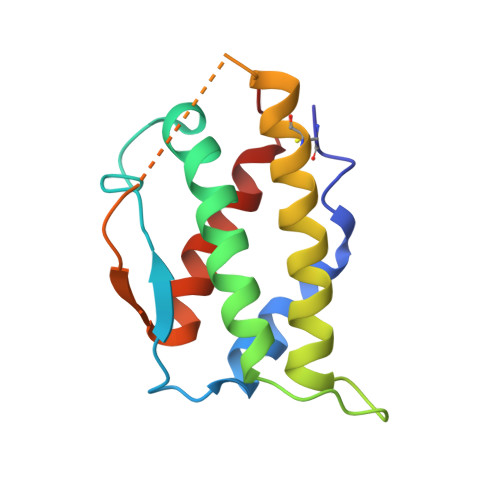
Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Krimmer, S.G., Bertoletti, N., Suzuki, Y., Katic, L., Mohanty, J., Shu, S., Lee, S., Lax, I., Mi, W., Schlessinger, J.(2023) Proc Natl Acad Sci U S A 120: e2300054120-e2300054120
- PubMed: 36943885
- DOI: https://doi.org/10.1073/pnas.2300054120
- Primary Citation of Related Structures:
8DFM, 8DFP, 8DFQ - PubMed Abstract:
The receptor tyrosine kinase KIT and its ligand stem cell factor (SCF) are required for the development of hematopoietic stem cells, germ cells, and other cells. A variety of human cancers, such as acute myeloid leukemia, gastrointestinal stromal tumor, and mast cell leukemia, are driven by somatic gain-of-function KIT mutations. Here, we report cryo electron microscopy (cryo-EM) structural analyses of full-length wild-type and two oncogenic KIT mutants, which show that the overall symmetric arrangement of the extracellular domain of ligand-occupied KIT dimers contains asymmetric D5 homotypic contacts juxtaposing the plasma membrane. Mutational analysis of KIT reveals in D5 region an "Achilles heel" for therapeutic intervention. A ligand-sensitized oncogenic KIT mutant exhibits a more comprehensive and stable D5 asymmetric conformation. A constitutively active ligand-independent oncogenic KIT mutant adopts a V-shaped conformation solely held by D5-mediated contacts. Binding of SCF to this mutant fully restores the conformation of wild-type KIT dimers, including the formation of salt bridges responsible for D4 homotypic contacts and other hallmarks of SCF-induced KIT dimerization. These experiments reveal an unexpected structural plasticity of oncogenic KIT mutants and a therapeutic target in D5.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520.
Organizational Affiliation: