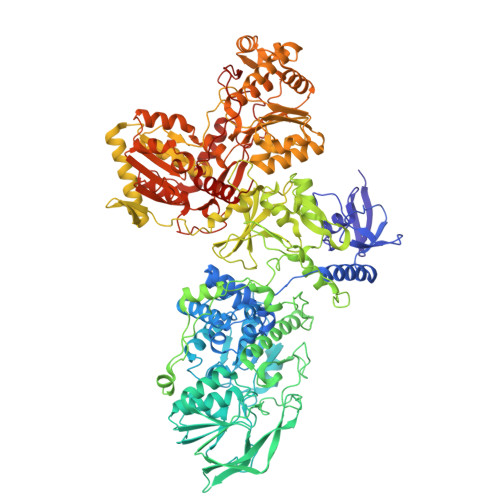
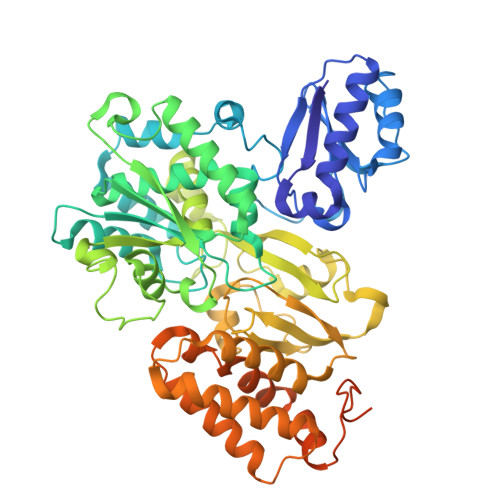
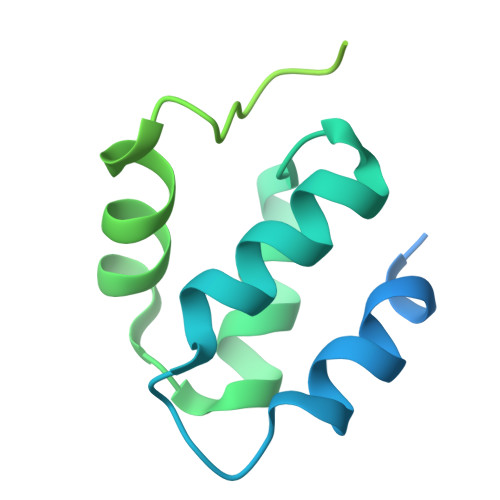
Cryo-EM reveals a composite flavobicluster electron bifurcation site in the Bfu family member NfnABC.
Li, H., Schut, G.J., Feng, X., Adams, M.W.W., Li, H.(2025) Commun Biol 8: 239-239
- PubMed: 39953182
- DOI: https://doi.org/10.1038/s42003-025-07706-8
- Primary Citation of Related Structures:
9BOV, 9BP5 - PubMed Abstract:
The BfuABC family is a diverse group of electron bifurcating enzymes that play key roles in anaerobic microbial metabolism. Previous studies have focused almost exclusively on the BfuABC-type hydrogenases but the mechanism and site of electron bifurcation remain unknown. Herein we focus on the Caldicellulosiruptor saccharolyticus (Csac) NfnABC-type Bfu enzyme that catalyzes the oxidation of NADPH and simultaneous reduction of NAD and the redox protein ferredoxin (Fd). Cryo-EM structures determined with and without NAD and Fd reveal seven FeS clusters and one FAD in NfnA, one FeS cluster in NfnC, and three FeS clusters, two Zn ions, and one FMN in NfnB. The Zn ions take the place of FeS clusters previously proposed in other Bfu family members. Csac Nfn for the first time defines the minimum bifurcation site as a flavobicluster consisting of FMN, a [4Fe-4S] (B1) cluster and a [2Fe-2S] (C1) cluster. Binding of NAD to the FMN triggers a series of conformational changes, crucial to the bifurcation of two electron pairs derived from NADPH by the [B1-FMN-C1] flavobicluster into low and high potential electrons that reduce Fd and NAD, respectively. The structures lay the foundation for investigations of the proposed reaction cycle common to all Bfu enzymes.
Organizational Affiliation:
Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.