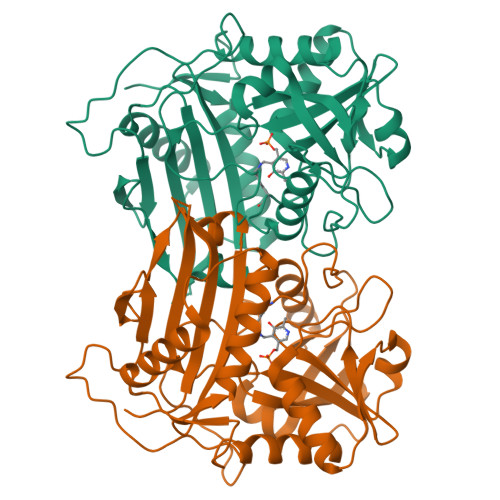
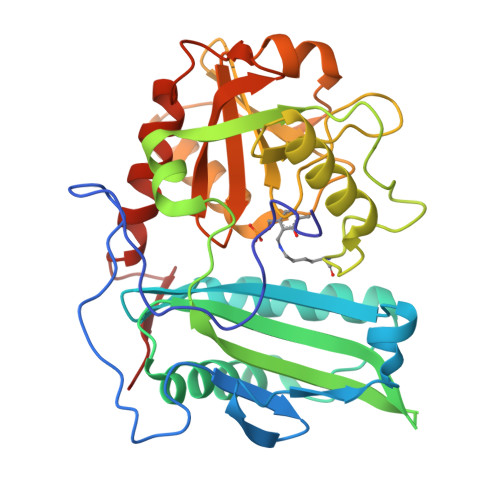
Structure-guided engineering an (R)-transaminase from Mycobacterium neoaurum for efficient synthesis of chiral N-heterocyclic amines.
Gao, X., Zhang, W., Wei, X., Zhao, L., Che, C., Zhang, Z., Wei, H., Qin, B., Liu, W., Jia, X., You, S.(2024) Int J Biol Macromol 287: 138591-138591
- PubMed: 39667461
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.138591
- Primary Citation of Related Structures:
9IJL, 9IVE - PubMed Abstract:
(R)-selective amine transaminases (R-ATAs) show considerable potential for the asymmetric synthesis of chiral drug intermediates. However, the low catalytic efficiency of natural R-ATAs toward bulky ketone substrates, such as N-heterocyclic compounds, severely limits its industrial application. In this study, five putative (R)-ATAs were mined from NCBI database, among which MnTA showed the highest activity for N-Boc-3-pyrrolidinone (1a) and N-Boc-3-piperidone (2a), and its crystal structure was performed. Furthermore, a structure-guided engineering strategy combined with directed evolution and in silico design was executed. Four key sites for substrate binding were identified based on alanine scanning. Then, a saturated mutation library was constructed, and residues G66 and F127 were found to be the key sites affecting substrate binding. By further combining mutation and iterative saturation mutation, variants with markedly improved activity were obtained. The optimal mutant MnTA-M1 (F127M) and MnTA-M5 (G66L/H67N/F127M/L160I) also displayed significantly enhanced activity toward various cyclic ketones or bulky N-heterocyclic ketone analogs. Finally, the gram-scale synthesis of (R)-3-amino-N-Boc-pyrrolidin (1b) and (R)-3-amino-N-Boc-piperidine (2b) was performed by the best mutants, achieving the space-time yields (STY) of 108 and 214 g/L·d, respectively. This research provides efficient biocatalysts for the synthesis of various chiral N-heterocyclic amines, along with a structural insight into the molecular mechanism for enhanced catalytic performance.
Organizational Affiliation:
School of Life Sciences and Biopharmaceutical Sciences, Shenyang Pharmaceutical University, 103 Wenhua Road, Shenhe, Shenyang 110016, People's Republic of China.