New Features
Search for Structural Motifs
02/16
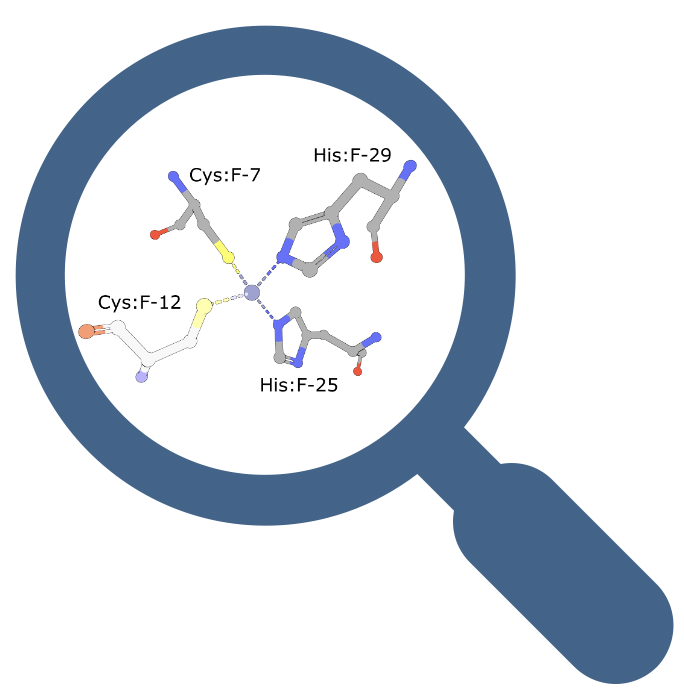
Structural motif similarities between PDB structures can provide valuable functional and evolutionary insights. A new Structural Motif service finds structures containing a small number of specific amino acids in proximity. This feature looks for residues that appear within 15 Å of each other; the residues may be located far apart in the sequence or appear in different polymer chains.
The search can be launched using the 3D viewer Mol* or through the Advanced Search>Structural Motif option. Help documentation is available.
This service rapidly searches the PDB using the functionality described in Real-time structural motif searching in proteins using an inverted index strategy 2020 PLoS Comput Biol 16(12): e1008502.